CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Polysaccharide Lyase Family 7"
| Line 44: | Line 44: | ||
== Three-dimensional structures == | == Three-dimensional structures == | ||
[[Image:EndovsexoPL7_Thomas_et_al_2013.jpg|thumb|400px|'''Figure 3. 3D Structure of endo- and exo-active PL7s''' <cite>Thomas2013</cite>. (A,B) endo AlyA1 and (C, D) exo AlyA5 from ''Zobellia galaactinovorans'' DsijT shown as cartoon (A,C) and surface structure (B,D) with superimposed tetrasaccharide from [{{PDBlink}}2ZAA PDB ID 2ZAA]. The image was conducted in PyMOL <cite>DeLano2002</cite>.]] | [[Image:EndovsexoPL7_Thomas_et_al_2013.jpg|thumb|400px|'''Figure 3. 3D Structure of endo- and exo-active PL7s''' <cite>Thomas2013</cite>. (A,B) endo AlyA1 and (C, D) exo AlyA5 from ''Zobellia galaactinovorans'' DsijT shown as cartoon (A,C) and surface structure (B,D) with superimposed tetrasaccharide from [{{PDBlink}}2ZAA PDB ID 2ZAA]. The image was conducted in PyMOL <cite>DeLano2002</cite>.]] | ||
| − | PL7 displays a jelly roll fold, which is also found in PL14 as well as in glycoside hydrolases of family [https://www.cazypedia.org/index.php/Glycoside_Hydrolase_Family_16 GH16]. ''Zobellia galactanivorans'' DsijT possess, among others, two PL7 with different modes of activity <cite>Thomas2013</cite>. AlyA1 is an endo-acting PL7 belonging to SF3 with a wide open cleft harboring the active site (Figure 3A, B), whereas AlyA5 belongs to SF5 and is exo-active which active site is closed by three loops forming a small pocket (Figure 3C, D). The highly conserved 9-amino-acid-block YFKAGVY*Q (where * is a variable residue) at the C-terminus of PL7s (Figure 2) has also been found for an extracellular pectate lyase (PL1, PDB: 1AIR) in ''E. chrysanthemi''. Alginate and pectate/pectin lyases share the β-elimination mechanism, the recognition of substrates of a similar structure, Ca<sup>2+</sup>-binding site and primary sequence similarity, indicating that they probably possess a similar core structural fold probably important to maintain a stable 3D-confirmation <cite>Wong2000</cite>. Several alginate lyases have been reported to be multimodular enzymes with putative non-catalytic, carbohydrate-binding modules (CBMs). The first biochemically characterized CBM of an endo PL7 was a N-terminal CBM13 from ''Agarivorans'' sp. L11, which increased its substrate binding and therefore catalytic efficiency, influenced substrate specificity, the product profile and thermo stability <cite>Li2015</cite>. Contradictory observations regarding catalytic efficiency and substrate specificity were made for an endo PL7 from ''Vibrio splendidus'' OU02 DNA with an N-terminal [https://www.cazypedia.org/index.php/Carbohydrate_Binding_Module_Family_32 CBM32] linked by a unique alpha-helix linker. The CBM and linker were proposed to serve as a "pivot point" somehow just pushing the product profile towards trisaccharides <cite>Lyu2018</cite>. The PL7 from ''Persicobacter'' sp. CCB-QB2 consists of three domains - a N-terminal [https://www.cazypedia.org/index.php/Carbohydrate_Binding_Module_Family_16 CBM16] with unknown function, a [https://www.cazypedia.org/index.php/Carbohydrate_Binding_Module_Family_32 CBM32] and the C-terminal PL7 <cite>Sim2017</cite>. The role of CBMs from different | + | PL7 displays a jelly roll fold, which is also found in PL14 as well as in glycoside hydrolases of family [https://www.cazypedia.org/index.php/Glycoside_Hydrolase_Family_16 GH16]. ''Zobellia galactanivorans'' DsijT possess, among others, two PL7 with different modes of activity <cite>Thomas2013</cite>. AlyA1 is an endo-acting PL7 belonging to SF3 with a wide open cleft harboring the active site (Figure 3A, B), whereas AlyA5 belongs to SF5 and is exo-active which active site is closed by three loops forming a small pocket (Figure 3C, D). The highly conserved 9-amino-acid-block YFKAGVY*Q (where * is a variable residue) at the C-terminus of PL7s (Figure 2) has also been found for an extracellular pectate lyase (PL1, PDB: 1AIR) in ''E. chrysanthemi''. Alginate and pectate/pectin lyases share the β-elimination mechanism, the recognition of substrates of a similar structure, Ca<sup>2+</sup>-binding site and primary sequence similarity, indicating that they probably possess a similar core structural fold probably important to maintain a stable 3D-confirmation <cite>Wong2000</cite>. |
| + | Several alginate lyases have been reported to be multimodular enzymes with putative non-catalytic, carbohydrate-binding modules (CBMs). The first biochemically characterized CBM of an endo PL7 was a N-terminal CBM13 from ''Agarivorans'' sp. L11, which increased its substrate binding and therefore catalytic efficiency, influenced substrate specificity, the product profile and thermo stability <cite>Li2015</cite>. Contradictory observations regarding catalytic efficiency and substrate specificity were made for an endo PL7 from ''Vibrio splendidus'' OU02 DNA with an N-terminal [https://www.cazypedia.org/index.php/Carbohydrate_Binding_Module_Family_32 CBM32] linked by a unique alpha-helix linker. The CBM and linker were proposed to serve as a "pivot point" somehow just pushing the product profile towards trisaccharides <cite>Lyu2018</cite>. The PL7 from ''Persicobacter'' sp. CCB-QB2 consists of three domains - a N-terminal [https://www.cazypedia.org/index.php/Carbohydrate_Binding_Module_Family_16 CBM16] with unknown function, a [https://www.cazypedia.org/index.php/Carbohydrate_Binding_Module_Family_32 CBM32] and the C-terminal PL7 <cite>Sim2017</cite>. The role of CBMs from different families as part of PL7s still remains unclear. | ||
| + | Flexible loops .... | ||
== Gene transfer of Alys among different habitats == | == Gene transfer of Alys among different habitats == | ||
Revision as of 03:11, 19 February 2020
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Authors: ^^^Nadine Gerlach^^^
- Responsible Curator: ^^^Jan-Hendrik Hehemann^^^
| Polysaccharide Lyase Family PL7 | |
| 3D Structure | β jelly roll |
| Mechanism | β-elimination |
| Active site residues | R, Q, H, 2xY |
| CAZy DB link | |
| http://www.cazy.org/PL7.html | |
Mechanism
Alginate lyases (Alys) of all families, PL5-7, PL14-15, PL17-18, catalyze the depolymerization of alginate in three steps: (I) removal of the negative charge on the carboxylate anion, (II) general base-catalyzed abstraction of the proton on the C5 and (III) β-elimination of the 4-O-glycosidic bond [2]. Most PL7s are endo-active, i.e. acting within a poly- or oligosaccharide and releasing smaller alginate fragments. Some PL7 are exo-acting cleaving a monosaccharide from the non-reducing end of the polymer [3]. In both modes of action, a new non-reducing end with a 4-deoxy-L-erythro-hex-4-en pyranosyl uronate residue (Δ) is formed.
Some marine PL7 alginate lyases require a calcium cation for substrate recognition and binding [3]. Ca2+ is weakening the ionic interactions between substrate (polyanion) and PL7 (polycation) by reducing the surface density of the alginate, charge and therefore increasing the enzyme activity [4]. However, there have also been reports where mono- or divalent (metal) cations did not increase or even decreased enzymatic activity [5, 6]. Therefore, it is not clear if Ca2+ is required for recognition and / or binding. However, one can speculate if Ca2+ is creating a more accessible form of alginate – the formation of gel particles, which might be an easier target in the marine environment as its dissolved form.
Kinetics and catalytic residues
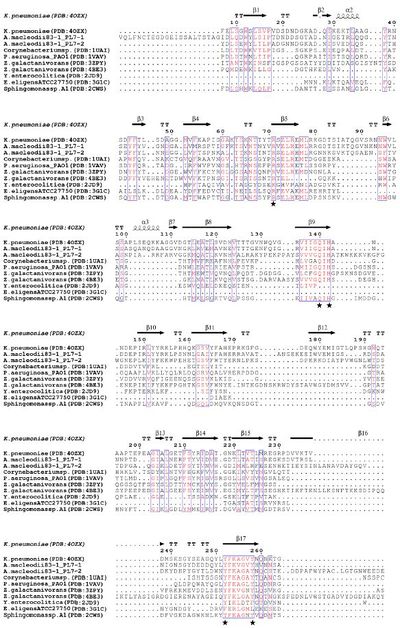
Several structural and biochemical analyses of wild type and mutated PL7s revealed five residues forming the active site: arginine (R), glutamine (Q), histidine (H), tyrosine (Y) [7, 8, 9], which are present in three highly conserved regions: R*ELR*ML, VIIGQ(I/V)H, YFKAG*Y*Q respectively (Figure 1) [10]. It has been proposed that in PL7 ALY-1 from Corynebacterium sp. Q117+Y195 interact near the reaction site of alginate to maintain proper orientation of the substrate, R72 interacts with alginate due to the formation of salt bridges with the carboxyl groups at the C5 and H119 acts as a base to deprotonate [11]. However, there can also be additional charged residues at the active site, which promote substrate recognition and binding [3]. Such residues can be found in the N-terminal R*ELREML and VIIGQIH regions. Both highly conserved regions are mainly characterized by hydrophobic amino acids (especially aromatic amino acids) such as leucine, tryptophan and methionine as well as residues with planar polar side chains (especially amino acids with charged side chains) such as arginine, glutamic acid, glutamine (Figure 2). These residues have been suggested to be substrate-binding molecules [10].
Substrate specificities
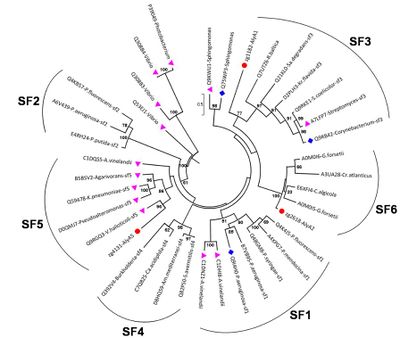
Polysaccharide lyase family 7 (PL7) contains five subfamilies (SF) based on their sequence similarities [12], plus a so far uncharacterized sixth subfamily, which consist only of marine representatives of the Flavobacteriaceae (Figure 2) [3]. The substrate specificity depends on the source of alginate, i.e. derived from brown seaweed or mucoid bacteria Pseudomonas spp. and Azotobacter vinelandii, as well as geographical and seasonal parameters. Alginate is a heteropolysaccharide, consisting of β-D-mannuronate (M) and α-L-guluronate (G). These monosaccharides can occur in homogenous and heterogenous blocks. In addition, bacterial alginate is often acetylated at the C2 and / or C3 of mannuronate, thereby shielding the substrate at this position for catalytic activity of Aly. Hence, PL7 lyases can be mannuronate (EC 4.2.2.3), guluronate (EC 4.2.2.11) or mixed link (EC 4.2.2.-) specific. Despite the preference for M- or G-enriched blocks, most PL7 also have a low to moderate activity for the other building block [3, 6, 13]. PolyG specific PL7 have been found in the SF3 and SF5 [3] with QIH in the second highly conserved region, while polyM specific PL7s are characterized by QVH [14, 15].
Three-dimensional structures
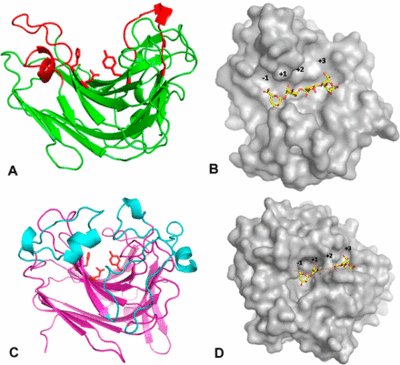
PL7 displays a jelly roll fold, which is also found in PL14 as well as in glycoside hydrolases of family GH16. Zobellia galactanivorans DsijT possess, among others, two PL7 with different modes of activity [3]. AlyA1 is an endo-acting PL7 belonging to SF3 with a wide open cleft harboring the active site (Figure 3A, B), whereas AlyA5 belongs to SF5 and is exo-active which active site is closed by three loops forming a small pocket (Figure 3C, D). The highly conserved 9-amino-acid-block YFKAGVY*Q (where * is a variable residue) at the C-terminus of PL7s (Figure 2) has also been found for an extracellular pectate lyase (PL1, PDB: 1AIR) in E. chrysanthemi. Alginate and pectate/pectin lyases share the β-elimination mechanism, the recognition of substrates of a similar structure, Ca2+-binding site and primary sequence similarity, indicating that they probably possess a similar core structural fold probably important to maintain a stable 3D-confirmation [10]. Several alginate lyases have been reported to be multimodular enzymes with putative non-catalytic, carbohydrate-binding modules (CBMs). The first biochemically characterized CBM of an endo PL7 was a N-terminal CBM13 from Agarivorans sp. L11, which increased its substrate binding and therefore catalytic efficiency, influenced substrate specificity, the product profile and thermo stability [17]. Contradictory observations regarding catalytic efficiency and substrate specificity were made for an endo PL7 from Vibrio splendidus OU02 DNA with an N-terminal CBM32 linked by a unique alpha-helix linker. The CBM and linker were proposed to serve as a "pivot point" somehow just pushing the product profile towards trisaccharides [18]. The PL7 from Persicobacter sp. CCB-QB2 consists of three domains - a N-terminal CBM16 with unknown function, a CBM32 and the C-terminal PL7 [13]. The role of CBMs from different families as part of PL7s still remains unclear. Flexible loops ....
Gene transfer of Alys among different habitats
Alginate degrading organisms often posse specified gene clusters for glycan utilization which contain, among other proteins such as transporters, several endo- and exo-acting Alys of different families. These gene clusters are called polysaccharide utilization loci (PULs) for Bacteriodetes [19] or alginolytic operons in case a SusCD pair transporter is missing or replaced by a different transporter system. It has been shown that these gene clusters can be transferred horizontally from one organism to another and thereby even cross different environmental habitats [20, 21]. The first alginate utilization system (AUS) was found in Zobellia galactinovorans which contains two clusters harboring five out of seven Alys (3x PL7). Those operons originated from an ancestral marine Flavobacterium and were independently transferred to marine Proteobacteria and Japanese gut Bacteriodetes by lateral gene transfer (LGT) [22].
A more detailed studied on horizontal gene transfer of PL7 between marine, ecophysiological different Vibrionaceae isolates revealed rapid adaptation of closely related but speciated bacterial populations, resulting into a fine scale of resource partitioning. The exchange of PL7 between marine microbes drove the evolution of polysaccharide degrading pathways, which might have led to three ecotypes – the pioneers, which degrade the polymer into oligomers, the harvester (intermediate of the other two types) and the scavenger, which can only utilize very small oligos created by the pioneers [23].
Family Firsts
- First catalytic endo-activity
- polyM PL7 from Photobacterium ATCC 433367 [24], polyG PL7 from Klebsiella pneumoniae subbsp. aerogenes [25]
- First catalytic exo-activity
- AlyA5 from Zobellia galactanivorans DsijT [3]
- First 3-D apo-structure
- PA1167 from Pseudomonas aeruginosa [26]
- First 3-D holo-structure
- A1-II from Sphingomons sp. A1 [27]
- First characterised CBM
- AlyL2 containing a N-terminal CBM13 from Agarivorans sp. L11 [17]
References
Error fetching PMID 23782694:
Error fetching PMID 22513138:
Error fetching PMID 20376150:
Error fetching PMID 15136569:
Error fetching PMID 25556193:
Error fetching PMID 16081095:
Error fetching PMID 24050202:
Error fetching PMID 25837818:
Error fetching PMID 25831216:
Error fetching PMID 11018131:
Error fetching PMID 29057942:
Error fetching PMID 29864445:
Error fetching PMID 20603004:
Error fetching PMID 8319887:
Error fetching PMID 8200539:
Error fetching PMID 15644208:
Error fetching PMID 24753421:
Error fetching PMID 11029455:
Error fetching PMID 24795372:
Error fetching PMID 27653556:
Error fetching PMID 29795267:
- Error fetching PMID 24753421:
-
Gacesa P. (1987) Alginate‐modifying enzymes: A proposed unified mechanism of action for the lyases and epimerases. FEBS Letters, 212, 1873-3468. DOI:10.1016/0014-5793(87)81344-3
- Error fetching PMID 23782694:
- Error fetching PMID 476128:
- Error fetching PMID 24795372:
- Error fetching PMID 25556193:
- Error fetching PMID 11029455:
- Error fetching PMID 16081095:
- Error fetching PMID 11018131:
- Error fetching PMID 15644208:
- Lombard V, Bernard T, Rancurel C, Brumer H, Coutinho PM, and Henrissat B. (2010). A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem J. 2010;432(3):437-44. DOI:10.1042/BJ20101185 |
- Error fetching PMID 29057942:
- Error fetching PMID 25831216:
- Error fetching PMID 24050202:
-
Delano WL. (2002) Pymol: An open-source molecular graphics tool. CCP4 Newsletter On Protein Crystallography, 40, 82-92. Direct link.
- Error fetching PMID 25837818:
- Error fetching PMID 29864445:
- Error fetching PMID 20376150:
- Error fetching PMID 29795267:
- Error fetching PMID 22513138:
- Error fetching PMID 27653556:
- Error fetching PMID 8319887:
- Error fetching PMID 8200539:
- Error fetching PMID 15136569:
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, and Henrissat B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(Database issue):D490-5. DOI:10.1093/nar/gkt1178 |
- Error fetching PMID 20603004: