CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Carbohydrate Binding Module Family 73
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| CAZy DB link | |
| https://www.cazy.org/CBM73.html |
Ligand specificities
Family 73 CBMs are modules of approximately 60 residues that are appended to bacterial enzymes associated to chitin degradation [1, 2, 3]. Binding to amorphous and crystalline α- and β-chitin has been demonstrated [1, 2]. The CBM73 module of the tri-modular LPMO, CjLPMO10A, binds tightly to α- chitin (Kd = 4.3 µM) and binding equilibrium was established within 15 minutes after mixing the protein and ligand [1].
Structural Features
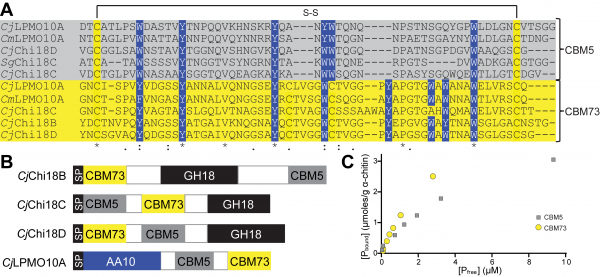
Currently no NMR or crystal structure is available for CBM73s, but circular dichroism experiments of the CBM from the Vibrio cholera GlcNAc-binding protein (GbpA, VcLPMO10B) indicated a β-sheet containing structure [2]. Sequence alignment shows that the CBM73 are distantly related to chitin-binding modules belonging to the CBM5 family (type A). Despite low sequence similarity, conserved aromatic amino acids within the CBM5 family, responsible for substrate-binding [4], align well with similar residues in the CBM73 family (Figure 1A)[1, 3]. Two additional aromatic residues are found in the CBM73 family compared to the CBM5 family. Compatibly, a somewhat lower Kd for α-chitin was observed for the C-terminal CBM73 of the Cellvibrio japonicus LPMO (CjLPMO10A) relative to its internal CBM5 (Figure 1BC)[1].
Functionalities
Family 73 CBMs are found in Gram-negative bacteria from the phylum of Proteobacteria and are covalently attached to chitin degrading enzymes such as GH18 and GH19 chitinases [2, 5], AA10 chitin-oxidizing lytic polysaccharide monooxygenases [1, 2] and often in combination with a CBM5 chitin-binding module. In chitin degrading enzymes from C. japonicus, the family 73 CBMs are found internally as well as at the N- or C-terminus (Figure 1B). The CBM73 from CjLPMO10A, together with the CBM5, strongly promotes targeting and binding of crystalline α- and β-chitin as the LPMO domain alone binds weakly to its substrate. Removal of the two CBMs (CBM5 and CBM73) in CjLPMO10A reduces the lifetime of the catalytic AA10 domain and decreases the overall product yield. A CBM73 has also been found appended to a serine protease/peptidoglycan hydrolase from Vibrio vulnificus. Truncation of the two CBMs (CBM5 and CBM73) resulted in reduced peptidoglycan hydrolyzing activity but did not affect the protease activity [6].
Family Firsts
- First Identified
- Family CBM73 was first found as a C-terminal module of the tri-modular Cellvibrio japonicus chitin-oxidizing LPMO (AA10-CBM5-CBM73) [1].
- First Structural Characterization
- Hitherto, no structural information is available
References
Error fetching PMID 27169553:
Error fetching PMID 16567413:
Error fetching PMID 18556790:
Error fetching PMID 21642466:
- Error fetching PMID 26858252:
- Wong E, Vaaje-Kolstad G, Ghosh A, Hurtado-Guerrero R, Konarev PV, Ibrahim AF, Svergun DI, Eijsink VG, Chatterjee NS, and van Aalten DM. (2012). The Vibrio cholerae colonization factor GbpA possesses a modular structure that governs binding to different host surfaces. PLoS Pathog. 2012;8(1):e1002373. DOI:10.1371/journal.ppat.1002373 |
- Error fetching PMID 27169553:
- Error fetching PMID 16567413:
- Error fetching PMID 18556790:
- Error fetching PMID 21642466: