CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Polysaccharide Lyase Family 17
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Polysaccharide Lyase Family 17 | |
| 3D structure | (α/α)6 barrel + anti-parallel β-sheet |
| Mechanism | β-elimination |
| Charge neutralizer | Asparagine and histidine |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/PL17.html | |
Substrate specificities
PL17 contains 2 subfamilies [1] as well as several proteins currently not assigned to any subfamily. Subfamily 2 has been shown to be exolytic alginate lyases [2, 3, 4, 5] with activity for all three block structures observed [6]. Alginate consisting of 1,4 linked β-D-mannuronic acid and α-L-guluronic acid arranged in poly-mannuronic acid , poly-guluronic acid or poly-mannuronic/guluronic acid blocks [7, 8]. Subfamily 1 has been found to be hyaluroran endo-lyases or poly-glucuronic acid lyases [6]. Hyaluronan consisting of N-acetyl-D-glucoamine and 1,4 linked D-glucoronic acid [9].
Kinetics and Mechanism
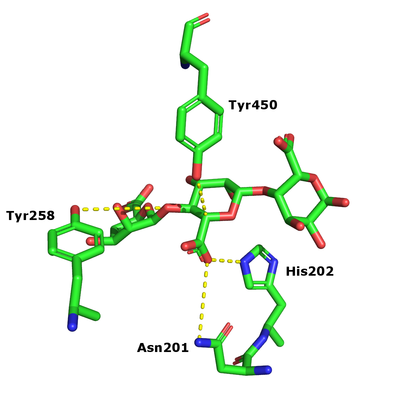
The β-elimination catalyzed by the PL17 enzymes results in the formation of a C4-C5 unsaturated sugar at the new non-reducing end. The first step is the neutralization of the acid group in the +1 subsite by the conserved histidine and asparagine. This lowers the pKa value of the C5-proton allowing for abstraction by the catalytic base (Figure 1). A catalytic acid then donates a proton to the glycosidic linkage resulting in the β-elimination [3].
Catalytic Residues
After charge neutralization a tyrosine functions as the catalytic base and another tyrosine as the acid. These were originally identified as Y456 and Y258 in Alg17c from Saccharophagus degradans [3].
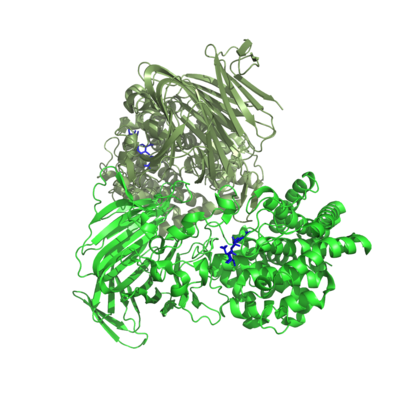
Three-dimensional structures
One crystal structure is available in PL17, that of Alg17c from Saccharophagus degradans belonging to subfamily 2 [3]. It is an (α/α)6 barrel + anti-parallel β-sheet with the catalytic machinery located in the (α/α)6 barrel (Figure 2). Alg17c is a homodimer, though that does not appear to be a general feature of PL17 [2, 3, 4, 5].
Family Firsts
- First catalytic activity
- MJ-3 alginate lyase assayed by monitoring the absorbance at 235 nm and characterizing the degradation products by TLC and 1H-NMR [10].
- First catalytic base/acid
- Y456 and Y258 in Alg17c crystal structure identified by their conservation in PL17, mutagenesis and kinetic analysis of mutants (Y258A and Y450A inactive) [3]
- First charge neutralizer
- N201 and H202 in the Alg17c crystal structure identified by their conservation in PL17, mutagenesis and kinetic analysis (N201A inactive and H202L 4.6 % activity remaining) [3]
- First 3-D structure
- Alg17c crystal structure [3]
References
Error fetching PMID 24795372:
Error fetching PMID 24478312:
Error fetching PMID 25335746:
Error fetching PMID 29795267:
Error fetching PMID 19870951:
Error fetching PMID 21826589:
- Error fetching PMID 20925655:
- Error fetching PMID 24795372:
- Error fetching PMID 24478312:
-
Shin, J. W., Lee, O. K., Park, H. H., Kim, H. S., and Lee, E. Y. (2015) Molecular characterization of a novel oligoalginate lyase consisting of AlgL- and heparinase II/III-like domains from Stenotrophomonas maltophilia KJ-2 and its application to alginate saccharification. Korean J. Chem. Eng. 32, 917–924 DOI:10.1007/s11814-014-0282-1
- Error fetching PMID 25335746:
- Error fetching PMID 29795267:
-
Haug, A., Larsen, B., and Smidsrod, O. (1967) Studies on sequence of uronic acid residues in alginic acid. Acta Chem. Scand. 21, 691–704 DOI:10.3891/acta.chem.scand.21-0691
-
Haug, A., Larsen, B., and Smidsrod, O. (1966) A study of constitution of alginic acid by partial acid hydrolysis. Acta Chem. Scand. 20, 183–190 DOI:10.3891/acta.chem.scand.20-0183
- Error fetching PMID 19870951:
- Error fetching PMID 21826589: