CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Difference between revisions of "Surface Binding Site"
Harry Brumer (talk | contribs) m (Text replacement - "\^\^\^(.*)\^\^\^" to "$1") |
|||
| (42 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
| − | {{ | + | {{CuratorApproved}} |
| − | * Authors: | + | * Authors: [[User:Birte Svensson|Birte Svensson]] and [[User:Darrell Cockburn|Darrell Cockburn]] |
| − | * Responsible Curators: | + | * Responsible Curators: [[User:Birte Svensson|Birte Svensson]] and [[User:Spencer Williams|Spencer Williams]] |
---- | ---- | ||
== Surface Binding Sites == | == Surface Binding Sites == | ||
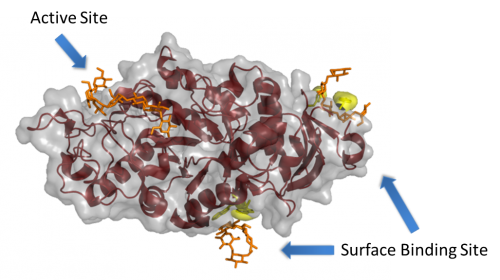
| − | [[Image:AMY1_SBS.png||thumb|right| | + | [[Image:AMY1_SBS.png||thumb|right|500px|'''Figure 1. The barley α-amylase 1 in complex with maltoheptaose PDB ID [{{PDBlink}}1rp8 1rp8]''' <cite>Robert2005</cite>. Several of the key SBS residues are shown highlighted in yellow, while the maltoheptaose molecules are shown in orange. Note the relatively large distance from the active site, which is a common aspect of these sites.]] |
| + | |||
A surface (or secondary) binding site (SBS) is a ligand binding site observed on the catalytic module of an enzyme, but outside of the active site itself (see Figure 1). For recent reviews on this topic, please see <cite>Cockburn2013 Cockburn2014 Cuyvers2012</cite>. | A surface (or secondary) binding site (SBS) is a ligand binding site observed on the catalytic module of an enzyme, but outside of the active site itself (see Figure 1). For recent reviews on this topic, please see <cite>Cockburn2013 Cockburn2014 Cuyvers2012</cite>. | ||
=== Detection and Occurrence === | === Detection and Occurrence === | ||
| − | SBSs have been observed in the crystal structures of approximately 50 carbohydrate active enzymes, with about half of these enzymes belonging to the [[GH13]] | + | SBSs have been observed in the crystal structures of approximately 50 carbohydrate active enzymes, with about half of these enzymes belonging to the family [[GH13]] (Table 1). Typically the enzymes found to possess one or more SBSs are active on polysaccharides, suggesting that SBSs are adaptations for dealing with longer substrates. X-ray crystallography has been the main method of detecting SBSs; however, NMR spectroscopy <cite>Ludwiczek2007</cite> and chemical labeling <cite>Gibson1987</cite> have also been used in the detection of these sites. Examination of the SBS containing enzymes show that they frequently co-occur with [[carbohydrate-binding modules]] (CBMs), suggesting that these two methods of binding to a substrate are complementary rather than redundant <cite>Cockburn2013</cite>. In one example, the α-amylase SusG from ''Bacteroides thetaiotaomicron'', both a CBM and an SBS were found to contribute to binding to starch granules <cite>Koropatkin2010</cite>. |
=== Roles of SBSs in Enzyme Function === | === Roles of SBSs in Enzyme Function === | ||
| − | Detailed analyses of SBSs have only been carried out in a few cases | + | Detailed analyses of SBSs have only been carried out in a few cases; however, in each of these cases they have been found to be important for the function of the enzyme. Various proven and speculated roles have been recently reviewed <cite>Cockburn2013 Cockburn2014 Cuyvers2012</cite>. In general the proposed roles of SBSs include: i) serving as an extension of the active site, guiding a substrate strand to the active site or maintaining binding to a polysaccharide strand to allow processivity, ii) acting as an allosteric regulator, with binding at the SBS affecting the properties of the active site, iii) serving as a pseudo-CBM, by targeting the enzyme to the substrate, anchoring the enzyme to the cell wall or disrupting the substrate (see the [[carbohydrate-binding modules]] page for more details on their functional roles). As an illustrative example, the two SBSs of the barley α-amylase 1 (named SBS1 and SBS2) <cite>Robert2005</cite> seem to fall into categories i) and iii). SBS1 is particularly important for the binding of the enzyme to starch granules <cite>Nielsen2009</cite>, while SBS2 is more important for the activity of the enzyme on amylopectin, lowering the apparent <i>K</i><sub>M</sub> for this substrate <cite>Nielsen2012</cite>. A good example of ii) is seen in the amylomaltase from ''Thermus aquaticus'', where binding to the SBS changes the active site, thereby altering the substrate profile of the enzyme <cite>Fugii2007</cite>. |
=== Studying SBSs === | === Studying SBSs === | ||
| − | The study of SBSs is often complicated by the presence | + | The study of SBSs is often complicated by the presence of multiple SBSs in a given catalytic module, substrate binding in the active site, or the presence of a CBM. Various techniques have been used to dissect contributions to SBSs such as the use of mutations, and substrates that do not bind at the active site <cite>Nielsen2009</cite> or the use of covalent inhibitors to block the active site <cite>Ludwiczek2007 Cuyvers2012b</cite>. A variety of techniques have proven useful for studying SBSs, including surface plasmon resonance, isothermal titration calorimetry, affinity electrophoresis and adsorption assays (the use of these techniques and others is summarized in <cite>Cockburn2013</cite>). |
| + | |||
| + | {| {{Prettytable}} width="400" | ||
| + | |- | ||
| + | |{{Hl2}} colspan="4" align="center"|'''Table 1: Glycoside hydrolase enzyme families for which an enzyme with an SBS has been identified.''' | ||
| + | |- | ||
| + | |'''Family''' | ||
| + | |'''# of Enzymes as of 2015-02-17''' | ||
| + | |'''Example Structure''' | ||
| + | |'''Reference(s)''' | ||
| + | |- | ||
| + | |[[GH1]]||2||[{{PDBlink}}1uyq 1uyq]||Unpublished | ||
| + | |- | ||
| + | |[[GH5]]||2||[{{PDBlink}}2pc8 2pc8]||<cite>Patrick2010</cite> | ||
| + | |- | ||
| + | |[[GH8]]||1||[{{PDBlink}}2b4f 2b4f]||<cite>DeVos2006</cite> | ||
| + | |- | ||
| + | |[[GH10]]||2||[{{PDBlink}}1goq 1goq]||<cite>LoLeggio2001 Schmidt1999</cite> | ||
| + | |- | ||
| + | |[[GH11]]||3||[{{PDBlink}}2qz3 2qz3]||<cite>Vandermarliere2008 Ludwiczek2007</cite> | ||
| + | |- | ||
| + | |[[GH13]]||24||[{{PDBlink}}1rp8 1rp8]||<cite>Robert2005 Cockburn2013 Cockburn2014</cite> | ||
| + | |- | ||
| + | |[[GH14]]||1||[{{PDBlink}}1b9z 1b9z]||<cite>Mikami1999</cite> | ||
| + | |- | ||
| + | |[[GH15]]||1||[{{PDBlink}}2f6d 2f6d]||<cite>Sevcik2006</cite> | ||
| + | |- | ||
| + | |[[GH16]]||1||[{{PDBlink}}1urx 1urx]||<cite>Allouch2004</cite> | ||
| + | |- | ||
| + | |[[GH19]]||1||[{{PDBlink}}3cql 3cql]||<cite>Huet2008</cite> | ||
| + | |- | ||
| + | |[[GH27]]||1||[{{PDBlink}}3hg2 3hg2]||<cite>Guce2010</cite> | ||
| + | |- | ||
| + | |[[GH31]]||1||[{{PDBlink}}3nqq 3nqq]||Unpublished | ||
| + | |- | ||
| + | |[[GH34]]||1||[{{PDBlink}}1mwe 1mwe]||<cite>Varghese1997</cite> | ||
| + | |- | ||
| + | |[[GH55]]||1||[{{PDBlink}}4pf0 4pf0]||<cite>Bianchetti2015</cite> | ||
| + | |- | ||
| + | |[[GH57]]||1||[{{PDBlink}}3n98 3n98]||<cite>Santos2010</cite> | ||
| + | |- | ||
| + | |[[GH63]]||1||[{{PDBlink}}3c67 3c67]||<cite>Kurakata2008</cite> | ||
| + | |- | ||
| + | |[[GH77]]||1||[{{PDBlink}}1esw 1esw]||<cite>Przylas2000</cite> | ||
| + | |- | ||
| + | |[[GH120]]||1||[{{PDBlink}}3vsv 3vsv]||<cite>Huang2012</cite> | ||
| + | |} | ||
== References == | == References == | ||
| Line 30: | Line 77: | ||
#Fugii2007 pmid=17368400 | #Fugii2007 pmid=17368400 | ||
#Cuyvers2012b pmid=21964501 | #Cuyvers2012b pmid=21964501 | ||
| − | + | #Patrick2010 pmid=20875088 | |
| + | #DeVos2006 pmid=16605248 | ||
| + | #LoLeggio2001 pmid=11741607 | ||
| + | #Schmidt1999 pmid=10029534 | ||
| + | #Vandermarliere2008 pmid=17983355 | ||
| + | #Mikami1999 pmid=10353816 | ||
| + | #Sevcik2006 pmid=16649993 | ||
| + | #Allouch2004 pmid=15062085 | ||
| + | #Huet2008 pmid=18636748 | ||
| + | #Guce2010 pmid=19940122 | ||
| + | #Varghese1997 pmid=9342319 | ||
| + | #Bianchetti2015 pmid=25752603 | ||
| + | #Santos2010 pmid=21104698 | ||
| + | #Kurakata2008 pmid=18586271 | ||
| + | #Przylas2000 pmid=11082203 | ||
| + | #Huang2012 pmid=22992047 | ||
</biblio> | </biblio> | ||
[[Category:Definitions and explanations]] | [[Category:Definitions and explanations]] | ||
Latest revision as of 13:17, 18 December 2021
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Authors: Birte Svensson and Darrell Cockburn
- Responsible Curators: Birte Svensson and Spencer Williams
Surface Binding Sites
A surface (or secondary) binding site (SBS) is a ligand binding site observed on the catalytic module of an enzyme, but outside of the active site itself (see Figure 1). For recent reviews on this topic, please see [2, 3, 4].
Detection and Occurrence
SBSs have been observed in the crystal structures of approximately 50 carbohydrate active enzymes, with about half of these enzymes belonging to the family GH13 (Table 1). Typically the enzymes found to possess one or more SBSs are active on polysaccharides, suggesting that SBSs are adaptations for dealing with longer substrates. X-ray crystallography has been the main method of detecting SBSs; however, NMR spectroscopy [5] and chemical labeling [6] have also been used in the detection of these sites. Examination of the SBS containing enzymes show that they frequently co-occur with carbohydrate-binding modules (CBMs), suggesting that these two methods of binding to a substrate are complementary rather than redundant [2]. In one example, the α-amylase SusG from Bacteroides thetaiotaomicron, both a CBM and an SBS were found to contribute to binding to starch granules [7].
Roles of SBSs in Enzyme Function
Detailed analyses of SBSs have only been carried out in a few cases; however, in each of these cases they have been found to be important for the function of the enzyme. Various proven and speculated roles have been recently reviewed [2, 3, 4]. In general the proposed roles of SBSs include: i) serving as an extension of the active site, guiding a substrate strand to the active site or maintaining binding to a polysaccharide strand to allow processivity, ii) acting as an allosteric regulator, with binding at the SBS affecting the properties of the active site, iii) serving as a pseudo-CBM, by targeting the enzyme to the substrate, anchoring the enzyme to the cell wall or disrupting the substrate (see the carbohydrate-binding modules page for more details on their functional roles). As an illustrative example, the two SBSs of the barley α-amylase 1 (named SBS1 and SBS2) [1] seem to fall into categories i) and iii). SBS1 is particularly important for the binding of the enzyme to starch granules [8], while SBS2 is more important for the activity of the enzyme on amylopectin, lowering the apparent KM for this substrate [9]. A good example of ii) is seen in the amylomaltase from Thermus aquaticus, where binding to the SBS changes the active site, thereby altering the substrate profile of the enzyme [10].
Studying SBSs
The study of SBSs is often complicated by the presence of multiple SBSs in a given catalytic module, substrate binding in the active site, or the presence of a CBM. Various techniques have been used to dissect contributions to SBSs such as the use of mutations, and substrates that do not bind at the active site [8] or the use of covalent inhibitors to block the active site [5, 11]. A variety of techniques have proven useful for studying SBSs, including surface plasmon resonance, isothermal titration calorimetry, affinity electrophoresis and adsorption assays (the use of these techniques and others is summarized in [2]).
| Table 1: Glycoside hydrolase enzyme families for which an enzyme with an SBS has been identified. | |||
| Family | # of Enzymes as of 2015-02-17 | Example Structure | Reference(s) |
| GH1 | 2 | 1uyq | Unpublished |
| GH5 | 2 | 2pc8 | [12] |
| GH8 | 1 | 2b4f | [13] |
| GH10 | 2 | 1goq | [14, 15] |
| GH11 | 3 | 2qz3 | [5, 16] |
| GH13 | 24 | 1rp8 | [1, 2, 3] |
| GH14 | 1 | 1b9z | [17] |
| GH15 | 1 | 2f6d | [18] |
| GH16 | 1 | 1urx | [19] |
| GH19 | 1 | 3cql | [20] |
| GH27 | 1 | 3hg2 | [21] |
| GH31 | 1 | 3nqq | Unpublished |
| GH34 | 1 | 1mwe | [22] |
| GH55 | 1 | 4pf0 | [23] |
| GH57 | 1 | 3n98 | [24] |
| GH63 | 1 | 3c67 | [25] |
| GH77 | 1 | 1esw | [26] |
| GH120 | 1 | 3vsv | [27] |
References
- Robert X, Haser R, Mori H, Svensson B, and Aghajari N. (2005). Oligosaccharide binding to barley alpha-amylase 1. J Biol Chem. 2005;280(38):32968-78. DOI:10.1074/jbc.M505515200 |
-
Cockburn, D. and Svensson, B. Surface binding sites in carbohydrate active enzymes: an emerging picture of structural and functional diversity. 2013. In: Lindhorst TK, Rauter AP (eds) SPR carbohydrate chemistry—chemical and biological approaches, vol 39. Royal Society of Chemistry, Cambridge. DOI: 10.1039/9781849737173-00204
-
Cockburn, D., Wilkens, C., Ruzanski, C., Andersen, S., Willum Nielsen, J., Smith, A.M., Field, R.A., Willemoës, M., Abou Hachem, M., and Svensson B. (2014) Analysis of surface binding sites (SBSs) in carbohydrate active enzymes with focus on glycoside hydrolase families 13 and 77 — a mini-review. Biologia, 69, 705-712. DOI: 10.2478/s11756-014-0373-9
- Cuyvers S, Dornez E, Delcour JA, and Courtin CM. (2012). Occurrence and functional significance of secondary carbohydrate binding sites in glycoside hydrolases. Crit Rev Biotechnol. 2012;32(2):93-107. DOI:10.3109/07388551.2011.561537 |
- Ludwiczek ML, Heller M, Kantner T, and McIntosh LP. (2007). A secondary xylan-binding site enhances the catalytic activity of a single-domain family 11 glycoside hydrolase. J Mol Biol. 2007;373(2):337-54. DOI:10.1016/j.jmb.2007.07.057 |
-
Gibson, RM, and Svensson, B. Identification of tryptophanyl residues involved in binding of carbohydrate ligands to barley α-amylase 2. Carlsberg Res Commun. 1987. 52: 373-379.
- Koropatkin NM and Smith TJ. (2010). SusG: a unique cell-membrane-associated alpha-amylase from a prominent human gut symbiont targets complex starch molecules. Structure. 2010;18(2):200-15. DOI:10.1016/j.str.2009.12.010 |
- Nielsen MM, Bozonnet S, Seo ES, Mótyán JA, Andersen JM, Dilokpimol A, Abou Hachem M, Gyémánt G, Naested H, Kandra L, Sigurskjold BW, and Svensson B. (2009). Two secondary carbohydrate binding sites on the surface of barley alpha-amylase 1 have distinct functions and display synergy in hydrolysis of starch granules. Biochemistry. 2009;48(32):7686-97. DOI:10.1021/bi900795a |
- Nielsen JW, Kramhøft B, Bozonnet S, Abou Hachem M, Stipp SL, Svensson B, and Willemoës M. (2012). Degradation of the starch components amylopectin and amylose by barley α-amylase 1: role of surface binding site 2. Arch Biochem Biophys. 2012;528(1):1-6. DOI:10.1016/j.abb.2012.08.005 |
- Fujii K, Minagawa H, Terada Y, Takaha T, Kuriki T, Shimada J, and Kaneko H. (2007). Function of second glucan binding site including tyrosines 54 and 101 in Thermus aquaticus amylomaltase. J Biosci Bioeng. 2007;103(2):167-73. DOI:10.1263/jbb.103.167 |
- Cuyvers S, Dornez E, Abou Hachem M, Svensson B, Hothorn M, Chory J, Delcour JA, and Courtin CM. (2012). Isothermal titration calorimetry and surface plasmon resonance allow quantifying substrate binding to different binding sites of Bacillus subtilis xylanase. Anal Biochem. 2012;420(1):90-2. DOI:10.1016/j.ab.2011.09.005 |
- Patrick WM, Nakatani Y, Cutfield SM, Sharpe ML, Ramsay RJ, and Cutfield JF. (2010). Carbohydrate binding sites in Candida albicans exo-β-1,3-glucanase and the role of the Phe-Phe 'clamp' at the active site entrance. FEBS J. 2010;277(21):4549-61. DOI:10.1111/j.1742-4658.2010.07869.x |
- De Vos D, Collins T, Nerinckx W, Savvides SN, Claeyssens M, Gerday C, Feller G, and Van Beeumen J. (2006). Oligosaccharide binding in family 8 glycosidases: crystal structures of active-site mutants of the beta-1,4-xylanase pXyl from Pseudoaltermonas haloplanktis TAH3a in complex with substrate and product. Biochemistry. 2006;45(15):4797-807. DOI:10.1021/bi052193e |
- Lo Leggio L, Kalogiannis S, Eckert K, Teixeira SC, Bhat MK, Andrei C, Pickersgill RW, and Larsen S. (2001). Substrate specificity and subsite mobility in T. aurantiacus xylanase 10A. FEBS Lett. 2001;509(2):303-8. DOI:10.1016/s0014-5793(01)03177-5 |
- Schmidt A, Gübitz GM, and Kratky C. (1999). Xylan binding subsite mapping in the xylanase from Penicillium simplicissimum using xylooligosaccharides as cryo-protectant. Biochemistry. 1999;38(8):2403-12. DOI:10.1021/bi982108l |
- Vandermarliere E, Bourgois TM, Rombouts S, Van Campenhout S, Volckaert G, Strelkov SV, Delcour JA, Rabijns A, and Courtin CM. (2008). Crystallographic analysis shows substrate binding at the -3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-beta-xylanases. Biochem J. 2008;410(1):71-9. DOI:10.1042/BJ20071128 |
- Mikami B, Adachi M, Kage T, Sarikaya E, Nanmori T, Shinke R, and Utsumi S. (1999). Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose. Biochemistry. 1999;38(22):7050-61. DOI:10.1021/bi9829377 |
- Sevcík J, Hostinová E, Solovicová A, Gasperík J, Dauter Z, and Wilson KS. (2006). Structure of the complex of a yeast glucoamylase with acarbose reveals the presence of a raw starch binding site on the catalytic domain. FEBS J. 2006;273(10):2161-71. DOI:10.1111/j.1742-4658.2006.05230.x |
- Allouch J, Helbert W, Henrissat B, and Czjzek M. (2004). Parallel substrate binding sites in a beta-agarase suggest a novel mode of action on double-helical agarose. Structure. 2004;12(4):623-32. DOI:10.1016/j.str.2004.02.020 |
- Huet J, Rucktooa P, Clantin B, Azarkan M, Looze Y, Villeret V, and Wintjens R. (2008). X-ray structure of papaya chitinase reveals the substrate binding mode of glycosyl hydrolase family 19 chitinases. Biochemistry. 2008;47(32):8283-91. DOI:10.1021/bi800655u |
- Guce AI, Clark NE, Salgado EN, Ivanen DR, Kulminskaya AA, Brumer H 3rd, and Garman SC. (2010). Catalytic mechanism of human alpha-galactosidase. J Biol Chem. 2010;285(6):3625-3632. DOI:10.1074/jbc.M109.060145 |
- Varghese JN, Colman PM, van Donkelaar A, Blick TJ, Sahasrabudhe A, and McKimm-Breschkin JL. (1997). Structural evidence for a second sialic acid binding site in avian influenza virus neuraminidases. Proc Natl Acad Sci U S A. 1997;94(22):11808-12. DOI:10.1073/pnas.94.22.11808 |
- Bianchetti CM, Takasuka TE, Deutsch S, Udell HS, Yik EJ, Bergeman LF, and Fox BG. (2015). Active site and laminarin binding in glycoside hydrolase family 55. J Biol Chem. 2015;290(19):11819-32. DOI:10.1074/jbc.M114.623579 |
- Santos CR, Tonoli CC, Trindade DM, Betzel C, Takata H, Kuriki T, Kanai T, Imanaka T, Arni RK, and Murakami MT. (2011). Structural basis for branching-enzyme activity of glycoside hydrolase family 57: structure and stability studies of a novel branching enzyme from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. Proteins. 2011;79(2):547-57. DOI:10.1002/prot.22902 |
- Kurakata Y, Uechi A, Yoshida H, Kamitori S, Sakano Y, Nishikawa A, and Tonozuka T. (2008). Structural insights into the substrate specificity and function of Escherichia coli K12 YgjK, a glucosidase belonging to the glycoside hydrolase family 63. J Mol Biol. 2008;381(1):116-28. DOI:10.1016/j.jmb.2008.05.061 |
- Przylas I, Terada Y, Fujii K, Takaha T, Saenger W, and Sträter N. (2000). X-ray structure of acarbose bound to amylomaltase from Thermus aquaticus. Implications for the synthesis of large cyclic glucans. Eur J Biochem. 2000;267(23):6903-13. DOI:10.1046/j.1432-1033.2000.01790.x |
- Huang CH, Sun Y, Ko TP, Chen CC, Zheng Y, Chan HC, Pang X, Wiegel J, Shao W, and Guo RT. (2012). The substrate/product-binding modes of a novel GH120 β-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485. Biochem J. 2012;448(3):401-7. DOI:10.1042/BJ20121359 |