CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Glycoside Hydrolase Family 103
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Author: ^^^Anthony Clarke^^^
- Responsible Curator: ^^^Anthony Clarke^^^
| Glycoside Hydrolase Family GH103 | |
| Clan | none
α+β "lysozyme fold" |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| http://www.cazy.org/fam/GH103.html | |
Substrate specificities
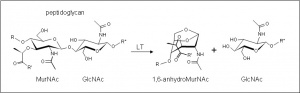
The glycoside hydrolases of this family are in fact lytic transglycosylases (also referred to as peptidoglycan lyases) of bacterial origin and they constitute family 3 of the classification scheme of Blackburn and Clarke [1]. The prototype for this family is membrane-bound lytic transglycosylase B (MltB) from Escherichia coli [2]. These enzymes cleave the β-1,4 linkage between N-acetylmuramoyl and N-acetylglucosaminyl residues in peptidoglycan (Figure 1). No other activities have been observed.
Kinetics and Mechanism
The lytic transglycosidases, strictly speaking, are retaining enzymes. However, they are not hydrolases but rather catalyse an intramolecular glycosyl transfer reaction onto the C-6 hydroxyl group of the muramoyl residue leading to the generation of a terminal 1,6-anhydromuramoyl product (Figure 1) that lacks a reducing end [3]. No detailed analyses involving both steady state and pre-steady state kinetic studies have been reported, but the Michaelis-Menten (KM and Vmax) parameters have been estimated for Pseudomonas aeruginosa MltB acting on insoluble peptidoglycan sacculi [4].
Catalytic Residues
As with other lytic transglycosylases (families GH23, GH102, and GH104), the GH103 enzymes are thought to possess a single catalytic general acid/base residue. This residue has been identified as Glu162 in MltB from both E. coli and P. aeruginosa and, indeed, its replacement abolishes catalytic activity [4, 5]. The mechanism of action of family GH103 enzymes has been investigated the most compared to the lytic transglycosylases of the other families (GH23,GH102, and GH104).
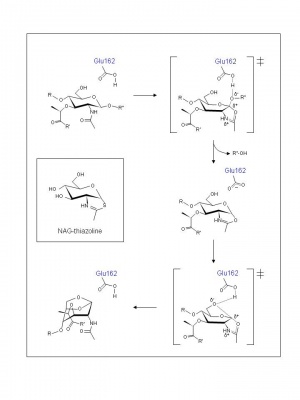
Examination of crystal structures of E. coli Slt35 (a soluble proteolytic derivative of MltB) and theoretical considerations led to the proposal of a mechanism that accommodates a single catalytic residue at its active site. Thus, based on the complexes formed with murodipeptide, chitobiose, and the inhibitor bulgecin, a two-step neighboring group participation mechanism involving substrate-assisted catalysis has been invoked analogous to the family GH18 chitinases and chitobiases, family GH20 N-acetyl-β-hexosaminidases, and family GH23 lytic transglycosylases [6]. Thus, the catalytic residue Glu162 is proposed to serve initially as a general acid to donate a proton to the glycosidic oxygen of the linkage to be cleaved. At the same time the MurNAc 2-acetamido group acts as a nucleophile and attacks the anomeric centre. The transition state leading to the intermediate possesses oxocarbenium ion character (Figure 2). In the second step abstraction of the C-6 hydroxyl proton of the oxazolinium species by Glu162 which now serves as a general base leads to nucleophilic attack and the formation of 1,6-anhydromuramic acid product, again through a transition state with oxocarbenium ion character. The β-hexosaminidase inhibitor NAG-thiazoline (Figure 2) was found to inhibit P. aeruginosa MltB thus supporting the proposal for the formation of an oxazolinium ion intermediate [7], and the results of a site-directed mutagenesis study suggest that Ser216 orients the N-acetyl group on MurNAc at the -1 subsite of MltB for its participation in a substrate-assisted mechanism of action [8].
Three-dimensional structures
Three-dimensional structures are available for several family GH103 enzymes, the first solved being that of E. coli MltB (Slt35) [5]. The catalytic domain of the enyzmes possesses the well characterized α+β "lysozyme fold."
Family Firsts
- First identification of lytic transglycosylase
- MltB from E. coli [2].
- First catalytic nucleophile identification
- Uses neighboring group participation mechanism.
- First general acid/base residue identification
- Inferred by X-ray crystallography of E. coli MltB [5].
- First 3-D structure
- E. coli MltB [5].
- First identification as a lipoprotein
- E. coli MltB [9].
- First identification of localization to outer membrane
- E. coli MltB [9].
- Frist demonstration of molecular interactions between GH103 enzymes and penicillin-binding proteins
- E. coli MltB [10].
References
- Blackburn NT and Clarke AJ. (2001). Identification of four families of peptidoglycan lytic transglycosylases. J Mol Evol. 2001;52(1):78-84. DOI:10.1007/s002390010136 |
- Engel H, Smink AJ, van Wijngaarden L, and Keck W. (1992). Murein-metabolizing enzymes from Escherichia coli: existence of a second lytic transglycosylase. J Bacteriol. 1992;174(20):6394-403. DOI:10.1128/jb.174.20.6394-6403.1992 |
- Höltje JV, Mirelman D, Sharon N, and Schwarz U. (1975). Novel type of murein transglycosylase in Escherichia coli. J Bacteriol. 1975;124(3):1067-76. DOI:10.1128/jb.124.3.1067-1076.1975 |
- Blackburn NT and Clarke AJ. (2002). Characterization of soluble and membrane-bound family 3 lytic transglycosylases from Pseudomonas aeruginosa. Biochemistry. 2002;41(3):1001-13. DOI:10.1021/bi011833k |
- van Asselt EJ, Dijkstra AJ, Kalk KH, Takacs B, Keck W, and Dijkstra BW. (1999). Crystal structure of Escherichia coli lytic transglycosylase Slt35 reveals a lysozyme-like catalytic domain with an EF-hand. Structure. 1999;7(10):1167-80. DOI:10.1016/s0969-2126(00)80051-9 |
- van Asselt EJ, Kalk KH, and Dijkstra BW. (2000). Crystallographic studies of the interactions of Escherichia coli lytic transglycosylase Slt35 with peptidoglycan. Biochemistry. 2000;39(8):1924-34. DOI:10.1021/bi992161p |
- Reid CW, Blackburn NT, Legaree BA, Auzanneau FI, and Clarke AJ. (2004). Inhibition of membrane-bound lytic transglycosylase B by NAG-thiazoline. FEBS Lett. 2004;574(1-3):73-9. DOI:10.1016/j.febslet.2004.08.006 |
- Reid CW, Legaree BA, and Clarke AJ. (2007). Role of Ser216 in the mechanism of action of membrane-bound lytic transglycosylase B: further evidence for substrate-assisted catalysis. FEBS Lett. 2007;581(25):4988-92. DOI:10.1016/j.febslet.2007.09.037 |
- Ehlert K, Höltje JV, and Templin MF. (1995). Cloning and expression of a murein hydrolase lipoprotein from Escherichia coli. Mol Microbiol. 1995;16(4):761-8. DOI:10.1111/j.1365-2958.1995.tb02437.x |
- von Rechenberg M, Ursinus A, and Höltje JV. (1996). Affinity chromatography as a means to study multienzyme complexes involved in murein synthesis. Microb Drug Resist. 1996;2(1):155-7. DOI:10.1089/mdr.1996.2.155 |