CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Carbohydrate Binding Module Family 9"
Harry Brumer (talk | contribs) m (fixed CBM Category tag for proper sorting) |
m |
||
| (25 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
<!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | <!-- RESPONSIBLE CURATORS: Please replace the {{UnderConstruction}} tag below with {{CuratorApproved}} when the page is ready for wider public consumption --> | ||
| − | {{ | + | {{CuratorApproved}} |
* [[Author]]: [[User:Johan Larsbrink|Johan Larsbrink]] | * [[Author]]: [[User:Johan Larsbrink|Johan Larsbrink]] | ||
* [[Responsible Curator]]: [[User:Johan Larsbrink|Johan Larsbrink]] | * [[Responsible Curator]]: [[User:Johan Larsbrink|Johan Larsbrink]] | ||
| Line 12: | Line 12: | ||
|{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | |{{Hl2}} colspan="2" align="center" |'''CAZy DB link''' | ||
|- | |- | ||
| − | | colspan="2" |{{CAZyDBlink}} | + | | colspan="2" |{{CAZyDBlink}}CBM9.html |
|} | |} | ||
</div> | </div> | ||
| Line 18: | Line 18: | ||
== Ligand specificities == | == Ligand specificities == | ||
| − | + | The tandem CBM9 domains found in the larger CBM22-CBM22-GH10-CBM9-CBM9 enzyme XynA from ''Thermotoga maritima'' (TmXynA) were initially shown to bind cellulose in pull-down studies <cite>Winterhalter1995</cite>. The (C-terminal) CBM9.2 domain was further studied using isothermal titration calorimetry (ITC), showing strongest binding to cellooligosaccharides but also weaker binding to lactose, maltose and xylobiose <cite>Boraston2001</cite>. Additionally, in depletion isotherms, the protein bound cellulose stronger than xylan. The CBM9 domains from the similar CBM22-GH10-CBM9-CBM9 XynX enzyme from ''Clostridium thermocellum'' was also suggested to bind cellulose <cite>Selvaraj2010</cite>. Later, a similar multidomain protein, ''Ck''Xyn10C-GE15A, from ''Caldicellulosiruptor kristjanssonii'' was studied and found to comprise a CBM22-CBM22-GH10-CBM9-CBM9-CBM9-CE15 architecture <cite>Krska2020</cite>. Its CBM9 domains (CBM9.1, CBM9.2 and CBM9.3) were shown to bind different glycans: in pull-down studies, CBM9.1 bound nothing tested, CBM9.2 bound cellulose, xylan, as well as mannan, and CBM9.3 bound cellulose and xylan though more weakly than CBM9.2. While using affinity gels, additional binding to xyloglucan was revealed for CBM9.3 <cite>Krska2021</cite>. This was also confirmed using ITC and differential scanning fluorometry where binding to xyloglucooligosaccharides was stronger than to cellooligosaccharides and xylooligosaccharides <cite>Krska2021</cite>. | |
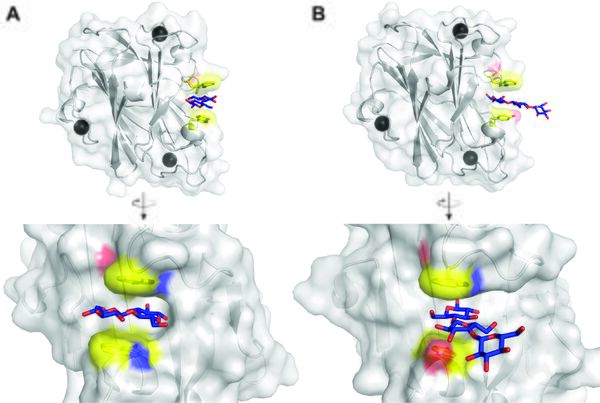
| − | + | [[File: Fig.1 CBM9.jpg|thumb|right|600px|'''Figure 1. CBM9 structures.''' '''A)''' Top: carton and surface representation of CBM9-2 from ''Thermotoga maritima'' ([{{PDBlink}}1i82 PDB 1i82]) <cite>Notenboom2001</cite>, with calcium ions as black spheres and cellobiose as blue sticks. Bottom: head-on view of the binding site in complex with cellobiose. '''B)''' Similar views as in A for CBM9.3 from ''Caldicellulosiruptor kristjanssonii'' with cellotriose as ligand ([{{PDBlink}}7nwq PDB 7nwq]) <cite>Krska2021</cite>.]] | |
| − | '' | ||
== Structural Features == | == Structural Features == | ||
| − | '' | + | The secondary structure of ''Tm''XynA CBM9.2 was initially shown to be mainly comprised of β-strands using circular dichroism <cite>Wassenberg1997</cite>, which was later confirmed when the structure was solved and showed a β-sandwich fold (Figure 1, [{{PDBlink}}1i8u PDB 1i8u]) <cite>Notenboom2001</cite>. The structure also revealed three calcium-binding sites, though not in close vicinity to the ligand binding site. A similar structure of the ''Ck''Xyn10C-GE15A CBM9.3 protein was later solved, again with bound calcium ions ([{{PDBlink}}7nwn PDB 7nwn]) <cite>Krska2021</cite>. The binding sites of both proteins differ, where that of ''Tm''CBM9.2 appears like a half-pocket, or blocked groove, able to accommodate two carbohydrate units, while that of ''Ck''CBM9.3 is a fully open groove. ''Tm''CBM9.2 was solved in complex with glucose ([{{PDBlink}}1i8A PDB 1i8a]) and cellobiose ([{{PDBlink}}1i82 PDB 1i82]) <cite>Notenboom2001</cite>), which revealed the cellobiose lying in the groove and being bound at the reducing end between two tryptophan residues. In ''Ck''CBM9.3 (solved separately with glucose ([{{PDBlink}}7nwo PDB 7nwo]), cellobiose ([{{PDBlink}}7nwp PDB 7nwp]), and cellotriose ([{{PDBlink}}7nwq PDB 7nwq]) <cite>Krska2021</cite>) the binding pose was however not aligned with the groove but the ligands found pointing perpendicular towards the protein and the reducing end bound between a tryptophan and a tyrosine residue. Curiously, cellotriose was bound simultaneously by two protomers facing each other, suggesting the possibility to bind either reducing- or non-reducing ends <cite>Krska2021</cite>. The binding type of characterized CBM9 proteins appears to be [[Carbohydrate-binding_modules#Types|type C]], binding chain ends, though the open groove of ''Ck''CBM9.3 suggests [[Carbohydrate-binding_modules#Types|type B]]-binders may exist in the family. |
| − | |||
| − | |||
| − | |||
== Functionalities == | == Functionalities == | ||
| − | + | CBM9 proteins are found almost exclusively in bacteria, with only a few eukaryotic and archaeal members in CAZy <cite>Drula2022</cite>. The majority of modules are found appended to enzymes related to xylan deconstruction, mainly GH10 xylanases, but also CEs from families 1, 4, 6, 15, and polyspecific families with potential xylanase activity such as GH5, 8, and 9. Also other functionalities such as putative agarase (GH50) or pectate lyase (PL9) domains are found as partners <cite>Drula2022</cite>, as well as DUFs <cite>Wong2017</cite>. Especially common are the CBM22-GH10-CBM9 motifs, with variable extensions of additional N-terminal CBM22 domains and C-terminal CBM9 domains as well as more catalytic modules on the same polypeptide, such as in ''Tm''XynA, ''Ct''XynX, and ''Ck''Xyn10C-GE15A <cite>Winterhalter1995 Selvaraj2010 Krska2020</cite>. | |
| − | + | ||
| − | + | CBM9 domains, while having been less studied than many other families, have been used as purification tags to enable cellulose-mediated protein affinity separation <cite>Kavoosi2004</cite>, and to increase protein thermostability <cite>Yang2018</cite>. | |
| − | |||
== Family Firsts == | == Family Firsts == | ||
| − | ;First Identified | + | ;First Identified: CBM9-2 from the larger XynA enzyme from ''Thermotoga maritima'' <cite>Winterhalter1995</cite>. |
| − | : | + | ;First Structural Characterization: CBM9-2 from the larger XynA enzyme from ''Thermotoga maritima'' <cite>Notenboom2001</cite> ([{{PDBlink}}1i8u PDB 1i8u]). |
| − | ;First Structural Characterization | ||
| − | : | ||
== References == | == References == | ||
<biblio> | <biblio> | ||
| − | # | + | #Winterhalter1995 pmid=7783614 |
| − | # | + | #Boraston2001 pmid=11371185 |
| − | # | + | #Selvaraj2010 pmid=21221946 |
| − | # | + | #Krska2020 pmid=32308737 |
| − | # | + | #Krska2021 pmid=34180241 |
| − | # | + | #Wassenberg1997 pmid=9260284 |
| − | # | + | #Notenboom2001 pmid=11371186 |
| + | #Drula2022 pmid=34850161 | ||
| + | #Wong2017 pmid=29326667 | ||
| + | #Kavoosi2004 pmid=15177165 | ||
| + | #Yang2018 pmid=29852224 | ||
</biblio> | </biblio> | ||
[[Category:Carbohydrate Binding Module Families|CBM009]] | [[Category:Carbohydrate Binding Module Families|CBM009]] | ||
Latest revision as of 04:37, 31 August 2023
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| CAZy DB link | |
| http://www.cazy.org/CBM9.html |
Ligand specificities
The tandem CBM9 domains found in the larger CBM22-CBM22-GH10-CBM9-CBM9 enzyme XynA from Thermotoga maritima (TmXynA) were initially shown to bind cellulose in pull-down studies [1]. The (C-terminal) CBM9.2 domain was further studied using isothermal titration calorimetry (ITC), showing strongest binding to cellooligosaccharides but also weaker binding to lactose, maltose and xylobiose [2]. Additionally, in depletion isotherms, the protein bound cellulose stronger than xylan. The CBM9 domains from the similar CBM22-GH10-CBM9-CBM9 XynX enzyme from Clostridium thermocellum was also suggested to bind cellulose [3]. Later, a similar multidomain protein, CkXyn10C-GE15A, from Caldicellulosiruptor kristjanssonii was studied and found to comprise a CBM22-CBM22-GH10-CBM9-CBM9-CBM9-CE15 architecture [4]. Its CBM9 domains (CBM9.1, CBM9.2 and CBM9.3) were shown to bind different glycans: in pull-down studies, CBM9.1 bound nothing tested, CBM9.2 bound cellulose, xylan, as well as mannan, and CBM9.3 bound cellulose and xylan though more weakly than CBM9.2. While using affinity gels, additional binding to xyloglucan was revealed for CBM9.3 [5]. This was also confirmed using ITC and differential scanning fluorometry where binding to xyloglucooligosaccharides was stronger than to cellooligosaccharides and xylooligosaccharides [5].
Structural Features
The secondary structure of TmXynA CBM9.2 was initially shown to be mainly comprised of β-strands using circular dichroism [7], which was later confirmed when the structure was solved and showed a β-sandwich fold (Figure 1, PDB 1i8u) [6]. The structure also revealed three calcium-binding sites, though not in close vicinity to the ligand binding site. A similar structure of the CkXyn10C-GE15A CBM9.3 protein was later solved, again with bound calcium ions (PDB 7nwn) [5]. The binding sites of both proteins differ, where that of TmCBM9.2 appears like a half-pocket, or blocked groove, able to accommodate two carbohydrate units, while that of CkCBM9.3 is a fully open groove. TmCBM9.2 was solved in complex with glucose (PDB 1i8a) and cellobiose (PDB 1i82) [6]), which revealed the cellobiose lying in the groove and being bound at the reducing end between two tryptophan residues. In CkCBM9.3 (solved separately with glucose (PDB 7nwo), cellobiose (PDB 7nwp), and cellotriose (PDB 7nwq) [5]) the binding pose was however not aligned with the groove but the ligands found pointing perpendicular towards the protein and the reducing end bound between a tryptophan and a tyrosine residue. Curiously, cellotriose was bound simultaneously by two protomers facing each other, suggesting the possibility to bind either reducing- or non-reducing ends [5]. The binding type of characterized CBM9 proteins appears to be type C, binding chain ends, though the open groove of CkCBM9.3 suggests type B-binders may exist in the family.
Functionalities
CBM9 proteins are found almost exclusively in bacteria, with only a few eukaryotic and archaeal members in CAZy [8]. The majority of modules are found appended to enzymes related to xylan deconstruction, mainly GH10 xylanases, but also CEs from families 1, 4, 6, 15, and polyspecific families with potential xylanase activity such as GH5, 8, and 9. Also other functionalities such as putative agarase (GH50) or pectate lyase (PL9) domains are found as partners [8], as well as DUFs [9]. Especially common are the CBM22-GH10-CBM9 motifs, with variable extensions of additional N-terminal CBM22 domains and C-terminal CBM9 domains as well as more catalytic modules on the same polypeptide, such as in TmXynA, CtXynX, and CkXyn10C-GE15A [1, 3, 4].
CBM9 domains, while having been less studied than many other families, have been used as purification tags to enable cellulose-mediated protein affinity separation [10], and to increase protein thermostability [11].
Family Firsts
- First Identified
- CBM9-2 from the larger XynA enzyme from Thermotoga maritima [1].
- First Structural Characterization
- CBM9-2 from the larger XynA enzyme from Thermotoga maritima [6] (PDB 1i8u).
References
- Winterhalter C, Heinrich P, Candussio A, Wich G, and Liebl W. (1995). Identification of a novel cellulose-binding domain within the multidomain 120 kDa xylanase XynA of the hyperthermophilic bacterium Thermotoga maritima. Mol Microbiol. 1995;15(3):431-44. DOI:10.1111/j.1365-2958.1995.tb02257.x |
- Boraston AB, Creagh AL, Alam MM, Kormos JM, Tomme P, Haynes CA, Warren RA, and Kilburn DG. (2001). Binding specificity and thermodynamics of a family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A. Biochemistry. 2001;40(21):6240-7. DOI:10.1021/bi0101695 |
- Selvaraj T, Kim SK, Kim YH, Jeong YS, Kim YJ, Phuong ND, Jung KH, Kim J, Yun HD, and Kim H. (2010). The role of carbohydrate-binding module (CBM) repeat of a multimodular xylanase (XynX) from Clostridium thermocellum in cellulose and xylan binding. J Microbiol. 2010;48(6):856-61. DOI:10.1007/s12275-010-0285-5 |
- Krska D and Larsbrink J. (2020). Investigation of a thermostable multi-domain xylanase-glucuronoyl esterase enzyme from Caldicellulosiruptor kristjanssonii incorporating multiple carbohydrate-binding modules. Biotechnol Biofuels. 2020;13:68. DOI:10.1186/s13068-020-01709-9 |
- Krska D, Mazurkewich S, Brown HA, Theibich Y, Poulsen JN, Morris AL, Koropatkin NM, Lo Leggio L, and Larsbrink J. (2021). Structural and Functional Analysis of a Multimodular Hyperthermostable Xylanase-Glucuronoyl Esterase from Caldicellulosiruptor kristjansonii. Biochemistry. 2021;60(27):2206-2220. DOI:10.1021/acs.biochem.1c00305 |
- Notenboom V, Boraston AB, Kilburn DG, and Rose DR. (2001). Crystal structures of the family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A in native and ligand-bound forms. Biochemistry. 2001;40(21):6248-56. DOI:10.1021/bi0101704 |
- Wassenberg D, Schurig H, Liebl W, and Jaenicke R. (1997). Xylanase XynA from the hyperthermophilic bacterium Thermotoga maritima: structure and stability of the recombinant enzyme and its isolated cellulose-binding domain. Protein Sci. 1997;6(8):1718-26. DOI:10.1002/pro.5560060812 |
- Drula E, Garron ML, Dogan S, Lombard V, Henrissat B, and Terrapon N. (2022). The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 2022;50(D1):D571-D577. DOI:10.1093/nar/gkab1045 |
- Wong MT, Wang W, Couturier M, Razeq FM, Lombard V, Lapebie P, Edwards EA, Terrapon N, Henrissat B, and Master ER. (2017). Comparative Metagenomics of Cellulose- and Poplar Hydrolysate-Degrading Microcosms from Gut Microflora of the Canadian Beaver (Castor canadensis) and North American Moose (Alces americanus) after Long-Term Enrichment. Front Microbiol. 2017;8:2504. DOI:10.3389/fmicb.2017.02504 |
- Kavoosi M, Meijer J, Kwan E, Creagh AL, Kilburn DG, and Haynes CA. (2004). Inexpensive one-step purification of polypeptides expressed in Escherichia coli as fusions with the family 9 carbohydrate-binding module of xylanase 10A from T. maritima. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;807(1):87-94. DOI:10.1016/j.jchromb.2004.03.031 |
- Yang A, Cheng J, Liu M, Shangguan Y, and Liu L. (2018). Sandwich fusion of CBM9_2 to enhance xylanase thermostability and activity. Int J Biol Macromol. 2018;117:586-591. DOI:10.1016/j.ijbiomac.2018.05.199 |