CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Carbohydrate Binding Module Family 80
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| CAZy DB link | |
| https://www.cazy.org/CBM80.html |
Ligand specificities
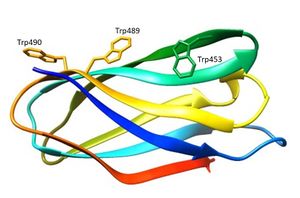
CBM80 is a small bacterial CBM family comprising around 96 amino acids and identified in the Ruminococcus flavefaciens cellulosome [1]. The only characterized CBM80 modules are CBM80RfGH5-1/2 and CBM80RfGH5, which are components of enzymes containing catalytic modules derived from GH5_4 (CAZy - GH5_4) and GH5_7 (CAZy - GH5_7) for CBM80RfGH5-1/2 and GH5_4 (CAZy - GH5_4) for CBM80RfGH5 [2]. CBM80RfGH5-1/2 and CBM80RfGH5 display specificity for β-1,4- and mixed linked β-1,3-1,4-glucans, while CBM80RfGH5-1/2 also binds β-1,4-mannans [2]. CBM80RfGH5-1/2 binds galactomannan in addition to the β-glucans with affinities in the range of 104 M-1 to 105 M-1 [2]. The endo-mode of binding to soluble polysaccharide indicates that CBM80 is a type B CBM [2]. The key residues implicated in CBM80RfGH5-1/2 ligand binding were identified by site-direct mutagenesis (Figure 1) [2]. Alanine substitution of Trp453 and Trp489 revealed a complete abrogation of binding to β-glucans and β-mannans, showing the importance of tryptophan residues in ligand recognition [2]. CBMs that bind to β-1,4-glycans typically contain three aromatic residues that make apolar interactions with sugars [3]. The predicted polar interactions between the protein and β-glycans have very little influence on affinity [2].
Structural Features
The three-dimensional structure of CBM80RfGH5-1/2 (5fu3) was solved using single-wavelength anomalous diffraction (SAD) methods and selenomethionyl labelled protein [2]. The structure of CBM80RfGH5-1/2 (Figure 1) apo, and in complex with mannohexaose and cellohexaose, was solved with resolutions of 1.0 Å, 1.4 Å and 1.5 Å, respectively [2]. CBM80RfGH5-1/2 has a β-sandwich fold consisting of two β-sheets (β-sheet 1 and 2) comprising of four anti-parallel β-strands each (Figure 1) [2]. The β-sheet 2 of CBM80RfGH-5-1/2 presents a planar hydrophobic surface with a parallel orientation of Trp453 and Trp489 and a perpendicular orientation of a third aromatic residue, Trp490 [2]. The mannohexaose CBM80RfGH5-1/2 complex revealed electron density for mannohexaose along the hydrophobic surface of β-sheet 2 [2]. The structure of CBM80RfGH5-1/2 in complex with cellohexaose revealed electron density for only three glucose units [2].
Functionalities
R. flavefaciens forms a multi-enzyme cellulosome complex that plays an integral role in the ability of this bacterium to degrade plant cell wall polysaccharides [4]. CBM80 modules play an enzyme-targeting role in a highly complex scaffold that is specific to the Ruminococcus [5]. CBM80RfGH5-1/2, a module in an enzyme that contains GH5_7 (CAZy - GH5_7) and GH5_4 catalytic modules (CAZy - GH5_4), binds β-glucans and β-mannans [2]. Other examples of CBMs that recognize both β-1,4-glucans and β-1,4-mannans are found in families CBM16 [6] and CBM29 [3]. CBM80RfGH5, a module in an enzyme that contains a GH5_4 (CAZy - GH5_4) catalytic module, displays specificity for β-glucans [2].
Family Firsts
- First Identified
- CBM80 from the Ruminococcus flavefaciens CBM80RfGH5-1/2 and CBM80RfGH5 [2].
- First Structural Characterization
- The first 3D crystal structure solved was CBM80RfGH5-1/2 [2].
References
- Rincon MT, Dassa B, Flint HJ, Travis AJ, Jindou S, Borovok I, Lamed R, Bayer EA, Henrissat B, Coutinho PM, Antonopoulos DA, Berg Miller ME, and White BA. (2010). Abundance and diversity of dockerin-containing proteins in the fiber-degrading rumen bacterium, Ruminococcus flavefaciens FD-1. PLoS One. 2010;5(8):e12476. DOI:10.1371/journal.pone.0012476 |
- Venditto I, Luis AS, Rydahl M, Schückel J, Fernandes VO, Vidal-Melgosa S, Bule P, Goyal A, Pires VM, Dourado CG, Ferreira LM, Coutinho PM, Henrissat B, Knox JP, Baslé A, Najmudin S, Gilbert HJ, Willats WG, and Fontes CM. (2016). Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition. Proc Natl Acad Sci U S A. 2016;113(26):7136-41. DOI:10.1073/pnas.1601558113 |
- Charnock SJ, Bolam DN, Nurizzo D, Szabó L, McKie VA, Gilbert HJ, and Davies GJ. (2002). Promiscuity in ligand-binding: The three-dimensional structure of a Piromyces carbohydrate-binding module, CBM29-2, in complex with cello- and mannohexaose. Proc Natl Acad Sci U S A. 2002;99(22):14077-82. DOI:10.1073/pnas.212516199 |
- Berg Miller ME, Antonopoulos DA, Rincon MT, Band M, Bari A, Akraiko T, Hernandez A, Thimmapuram J, Henrissat B, Coutinho PM, Borovok I, Jindou S, Lamed R, Flint HJ, Bayer EA, and White BA. (2009). Diversity and strain specificity of plant cell wall degrading enzymes revealed by the draft genome of Ruminococcus flavefaciens FD-1. PLoS One. 2009;4(8):e6650. DOI:10.1371/journal.pone.0006650 |
- Bensoussan L, Moraïs S, Dassa B, Friedman N, Henrissat B, Lombard V, Bayer EA, and Mizrahi I. (2017). Broad phylogeny and functionality of cellulosomal components in the bovine rumen microbiome. Environ Microbiol. 2017;19(1):185-197. DOI:10.1111/1462-2920.13561 |
- Bae B, Ohene-Adjei S, Kocherginskaya S, Mackie RI, Spies MA, Cann IK, and Nair SK. (2008). Molecular basis for the selectivity and specificity of ligand recognition by the family 16 carbohydrate-binding modules from Thermoanaerobacterium polysaccharolyticum ManA. J Biol Chem. 2008;283(18):12415-25. DOI:10.1074/jbc.M706513200 |