CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Glycoside Hydrolase Family 16
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Glycoside Hydrolase Family 16 | |
| Clan | GH-B |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/GH16.html | |
Substrate specificities
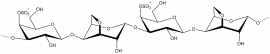
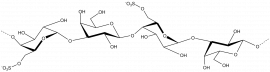
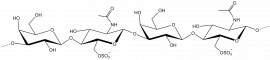
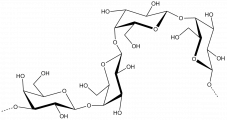
The members of family 16 are active on β-1,4 or β-1,3 glycosidic bonds in various glucans and galactans. A wide diversity of glycoside hydrolases active on plant and marine polysaccharides are found in GH16, including:
- keratan-sulfate endo-1,4-β-galactosidases (EC 3.2.1.103),
- endo-1,3-β-galactanases (EC 3.2.1.-),
- endo-1,3-β-glucanases (EC 3.2.1.39),
- endo-1,3(4)-β-glucanases (EC 3.2.1.6),
- licheninases (EC 3.2.1.73),
- β-agarases (EC 3.2.1.81),
- β-porphyranases (EC 3.2.1.178) [1],
- κ-carrageenases (EC 3.2.1.83), and
- endo-xyloglucanases (EC 3.2.1.151, a.k.a. xyloglucan endo-hydrolases, XEHs, in plants [2]).
Notably, some members of GH16 are predominant transglycosylases. These include the plant xyloglucan:xyloglucosyltransferases (EC 2.4.1.207, a.k.a. xyloglucan endo-transglycosylases, XETs) [2] and yeast chitin/beta-glucan crosslinking enzymes Crh1 and Crh2 [3, 4, 5]. Some invertebrate GH16 proteins have lost their catalytic amino acids and are involved in immune response activation through the Toll pathway upon binding of β-1,3 glucan. The role of the GH16 domain in this immune response has not been fully elucidated [6].
Several of the activities observed for GH16 members are delineated into individual sequence-based subfamilies, while other polyspecific subfamilies capture a range of activities [7].
- Diverse polysaccharides cleaved by GH16 enzymes
Kinetics and Mechanism
Members of GH16 enzymes are retaining enzymes, as first shown by NMR [8] on an endo-1,3-1,4-β-D-glucan 4-glucanohydrolase from Bacillus licheniformis. As such, they utilize a covalent glycosyl-enzyme intermediate, which is broken-down by glycosyl transfer [9, 10] to water or a carbohydrate acceptor substrate in glycoside hydrolases or transglycosylases, respectively.
Catalytic Residues
The catalytic nucleophile of GH16 enzymes was first proposed using a non-specific epoxyalkyl β-glycoside inhibitor and identification of the site of covalent labelling using ESI-MS and Edman degradation on an endo-1,3-1,4-β-D-glucan 4-glucanohydrolase from Bacillus amyloliquefaciens [11]. This was subsequently verified by azide rescue of the E134A mutant of a Bacillus licheniformis 1,3-1,4-β-D-glucan 4-glucanohydrolase resulting in an α-glycosyl azide from the β-glycoside substrate [12]. The general acid/base residue was identified by making the E138A site-directed mutant of the Bacillus licheniformis 1,3-1,4-β-D-glucan 4-glucanohydrolase together with kinetic analysis and azide rescue, which resulted in a β-glycosyl azide product [12]. These structurally conserved catalytic residues have been confirmed in a number of other GH16 members, including plant XETs and XEHs [13, 14], and yeast Crh1 and Crh2 [5].
The mechanistic analysis of bacterial mixed-linkage endo-glucanases has been expertly reviewed in the broader context of GH16 [15].
Three-dimensional structures
Proteins in GH16 share a β-jelly-roll fold in which two β-sheets align in a curved, sandwich-like manner and present a cleft-shaped active-site bounded by loops extending from the β-strands. The first solved 3D structure was a hybrid protein of licheninase M from Paenibacillus macerans and BglA from Bacillus amyloliquefaciens (PDB 1byh) in 1992 [16]. Many three-dimensional structures have been solved of family 16 members of archeal, bacterial, and eukaryotic origin (see http://www.cazy.org/GH16_structure.html for an updated list). Of these, the first eukaryotic 3D structure was the xyloglucan endo-transglycosylase PttXET16-34 from Populus tremula×tremuloides (PDB 1umz) [17] and the first archeal 3D structure was a endo-1,3-β-glucanase Lam16 from Pyrococcus furiosus (PDB 2vy0) [18].
The structural diversity of GH16 members across sequence-related subfamilies has been reviewed in detail [7].
Evolution of GH16
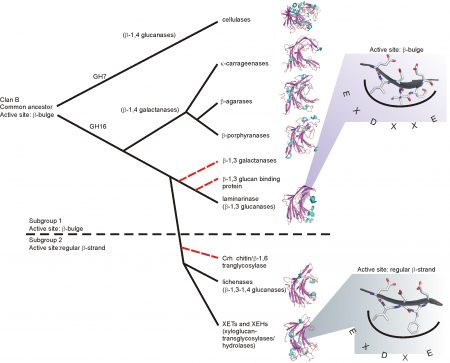
GH16 is a member of clan GH-B together with GH7; both families share the β-jellyroll fold. The different specificities of GH16 are proposed to have evolved from an ancestral β-1,3-glucanase [19]. This proposal was first elaborated using a structure-based phylogeny approach, which suggested that an early branching event lead to the evolution of the bacterial κ-carrageenases and the β-agarases, while a later branching event lead to the bacterial licheninases and the plant XETs [20] (Figure 1). GH16 has more recently been divided into subfamilies within CAZy, the corresponding phylogenetic analysis of which supports this overall evolutionary trajectory [7].
Particularly notable, the GH16 active-site residues are located in-train on one beta-strand at the center of the substrate binding cleft. Depending upon the phylogenetic clade, this beta-strand features one of two topologies. The beta-bulge motif, which has the consensus sequence EXDXXE, is more frequent in GH16 compared to the regular beta-strand with the consensus sequence EXDXE (the catalytic nucleophile is the first glutamate and the catalytic acid/base is the second, with a proposed "helper" asparate in-between [15]). Due to the predominance of the beta-bulge motif and its presence as the only motif in GH7, Michel et al. proposed that the beta-bulge is the ancestral motif, which subsequently gave rise to the regular beta-strand of extant plant XETs and bacterial licheninases [20].
Within plant lineages, similar structure-based phylogenetic approaches have suggested that XEHs evolved subsequently to XEHs within the xyloglucan endo-transglycosylase/hydrolase (XTH) gene family [2, 21]. The identification of a group of bifunctional GH16 glycoside hydrolases, which is active on both mixed-linkage beta-glucan and xyloglucan, provides additional support for the close evolutionary relationship of XETs and licheninases [22, 23, 24].
Family firsts
- First stereochemistry determination
- Bacillus licheniformis 1,3-1,4-β-D-glucan 4-glucanohydrolase by NMR [8].
- First catalytic nucleophile identification
- Suggested in Bacillus amyloliquefaciens 1,3-1,4-β-D-glucan 4-glucanohydrolase via non-specific epoxyalkyl β-glycoside labeling [11]. Later verified by azide rescue of inactivated mutants [12].
- First general acid/base residue identification
- Bacillus licheniformis 1,3-1,4-β-D-glucan 4-glucanohydrolase, first suggested by sequence homology and mutational studies [25]. This was later verified by azide rescue of inactivated mutants [12].
- First 3-D structure
- A hybrid licheninase (Bacillus amyloliquefaciens and Paenibacillus macerans) by X-ray crystallography (PDB 1byh) [16].
Reference list
- Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, and Michel G. (2010). Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464(7290):908-12. DOI:10.1038/nature08937 |
- Eklöf JM and Brumer H. (2010). The XTH gene family: an update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol. 2010;153(2):456-66. DOI:10.1104/pp.110.156844 |
- Cabib E, Farkas V, Kosík O, Blanco N, Arroyo J, and McPhie P. (2008). Assembly of the yeast cell wall. Crh1p and Crh2p act as transglycosylases in vivo and in vitro. J Biol Chem. 2008;283(44):29859-72. DOI:10.1074/jbc.M804274200 |
- Mazáň M, Blanco N, Kováčová K, Firáková Z, Rehulka P, Farkaš V, and Arroyo J. (2013). A novel fluorescence assay and catalytic properties of Crh1 and Crh2 yeast cell wall transglycosylases. Biochem J. 2013;455(3):307-18. DOI:10.1042/BJ20130354 |
- Blanco N, Sanz AB, Rodríguez-Peña JM, Nombela C, Farkaš V, Hurtado-Guerrero R, and Arroyo J. (2015). Structural and functional analysis of yeast Crh1 and Crh2 transglycosylases. FEBS J. 2015;282(4):715-31. DOI:10.1111/febs.13176 |
- Lee H, Kwon HM, Park JW, Kurokawa K, and Lee BL. (2009). N-terminal GNBP homology domain of Gram-negative binding protein 3 functions as a beta-1,3-glucan binding motif in Tenebrio molitor. BMB Rep. 2009;42(8):506-10. DOI:10.5483/bmbrep.2009.42.8.506 |
- Viborg AH, Terrapon N, Lombard V, Michel G, Czjzek M, Henrissat B, and Brumer H. (2019). A subfamily roadmap of the evolutionarily diverse glycoside hydrolase family 16 (GH16). J Biol Chem. 2019;294(44):15973-15986. DOI:10.1074/jbc.RA119.010619 |
- Malet C, Jiménez-Barbero J, Bernabé M, Brosa C, and Planas A. (1993). Stereochemical course and structure of the products of the enzymic action of endo-1,3-1,4-beta-D-glucan 4-glucanohydrolase from Bacillus licheniformis. Biochem J. 1993;296 ( Pt 3)(Pt 3):753-8. DOI:10.1042/bj2960753 |
-
Sinnott, M.L. (1990) Catalytic mechanisms of enzymic glycosyl transfer. Chem. Rev. 90, 1171-1202. DOI: 10.1021/cr00105a006
- Bissaro B, Monsan P, Fauré R, and O'Donohue MJ. (2015). Glycosynthesis in a waterworld: new insight into the molecular basis of transglycosylation in retaining glycoside hydrolases. Biochem J. 2015;467(1):17-35. DOI:10.1042/BJ20141412 |
- Høj PB, Condron R, Traeger JC, McAuliffe JC, and Stone BA. (1992). Identification of glutamic acid 105 at the active site of Bacillus amyloliquefaciens 1,3-1,4-beta-D-glucan 4-glucanohydrolase using epoxide-based inhibitors. J Biol Chem. 1992;267(35):25059-66. | Google Books | Open Library
- Viladot JL, de Ramon E, Durany O, and Planas A. (1998). Probing the mechanism of Bacillus 1,3-1,4-beta-D-glucan 4-glucanohydrolases by chemical rescue of inactive mutants at catalytically essential residues. Biochemistry. 1998;37(32):11332-42. DOI:10.1021/bi980586q |
- Gullfot F, Ibatullin FM, Sundqvist G, Davies GJ, and Brumer H. (2009). Functional characterization of xyloglucan glycosynthases from GH7, GH12, and GH16 scaffolds. Biomacromolecules. 2009;10(7):1782-8. DOI:10.1021/bm900215p |
- Piens K, Henriksson AM, Gullfot F, Lopez M, Fauré R, Ibatullin FM, Teeri TT, Driguez H, and Brumer H. (2007). Glycosynthase activity of hybrid aspen xyloglucan endo-transglycosylase PttXET16-34 nucleophile mutants. Org Biomol Chem. 2007;5(24):3971-8. DOI:10.1039/b714570e |
- Planas A (2000). Bacterial 1,3-1,4-beta-glucanases: structure, function and protein engineering. Biochim Biophys Acta. 2000;1543(2):361-382. DOI:10.1016/s0167-4838(00)00231-4 |
- Keitel T, Simon O, Borriss R, and Heinemann U. (1993). Molecular and active-site structure of a Bacillus 1,3-1,4-beta-glucanase. Proc Natl Acad Sci U S A. 1993;90(11):5287-91. DOI:10.1073/pnas.90.11.5287 |
- Johansson P, Brumer H 3rd, Baumann MJ, Kallas AM, Henriksson H, Denman SE, Teeri TT, and Jones TA. (2004). Crystal structures of a poplar xyloglucan endotransglycosylase reveal details of transglycosylation acceptor binding. Plant Cell. 2004;16(4):874-86. DOI:10.1105/tpc.020065 |
- Ilari A, Fiorillo A, Angelaccio S, Florio R, Chiaraluce R, van der Oost J, and Consalvi V. (2009). Crystal structure of a family 16 endoglucanase from the hyperthermophile Pyrococcus furiosus--structural basis of substrate recognition. FEBS J. 2009;276(4):1048-58. DOI:10.1111/j.1742-4658.2008.06848.x |
- Barbeyron T, Gerard A, Potin P, Henrissat B, and Kloareg B. (1998). The kappa-carrageenase of the marine bacterium Cytophaga drobachiensis. Structural and phylogenetic relationships within family-16 glycoside hydrolases. Mol Biol Evol. 1998;15(5):528-37. DOI:10.1093/oxfordjournals.molbev.a025952 |
- Michel G, Chantalat L, Duee E, Barbeyron T, Henrissat B, Kloareg B, and Dideberg O. (2001). The kappa-carrageenase of P. carrageenovora features a tunnel-shaped active site: a novel insight in the evolution of Clan-B glycoside hydrolases. Structure. 2001;9(6):513-25. DOI:10.1016/s0969-2126(01)00612-8 |
- Baumann MJ, Eklöf JM, Michel G, Kallas AM, Teeri TT, Czjzek M, and Brumer H 3rd. (2007). Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: biological implications for cell wall metabolism. Plant Cell. 2007;19(6):1947-63. DOI:10.1105/tpc.107.051391 |
- Eklöf JM, Shojania S, Okon M, McIntosh LP, and Brumer H. (2013). Structure-function analysis of a broad specificity Populus trichocarpa endo-β-glucanase reveals an evolutionary link between bacterial licheninases and plant XTH gene products. J Biol Chem. 2013;288(22):15786-99. DOI:10.1074/jbc.M113.462887 |
- McGregor N, Yin V, Tung CC, Van Petegem F, and Brumer H. (2017). Crystallographic insight into the evolutionary origins of xyloglucan endotransglycosylases and endohydrolases. Plant J. 2017;89(4):651-670. DOI:10.1111/tpj.13421 |
- Behar H, Graham SW, and Brumer H. (2018). Comprehensive cross-genome survey and phylogeny of glycoside hydrolase family 16 members reveals the evolutionary origin of EG16 and XTH proteins in plant lineages. Plant J. 2018;95(6):1114-1128. DOI:10.1111/tpj.14004 |
- Juncosa M, Pons J, Dot T, Querol E, and Planas A. (1994). Identification of active site carboxylic residues in Bacillus licheniformis 1,3-1,4-beta-D-glucan 4-glucanohydrolase by site-directed mutagenesis. J Biol Chem. 1994;269(20):14530-5. | Google Books | Open Library
- Kotake T, Hirata N, Degi Y, Ishiguro M, Kitazawa K, Takata R, Ichinose H, Kaneko S, Igarashi K, Samejima M, and Tsumuraya Y. (2011). Endo-beta-1,3-galactanase from winter mushroom Flammulina velutipes. J Biol Chem. 2011;286(31):27848-54. DOI:10.1074/jbc.M111.251736 |