CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Glycoside Hydrolase Family 46
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Glycoside Hydrolase Family GH46 | |
| Clan | GH-I |
| Mechanism | inverting |
| Active site residues | known |
| CAZy DB link | |
| https://www.cazy.org/GH46.html | |
Substrate specificities
Glycoside hydrolases of family 46 are essentially all endo-β-1,4-chitosanases (EC 3.2.1.132) that hydrolyze various links in chitosan, a polymer of β-1,4-linked D-glucosamine (GlcN) units with a variable content (mostly 0 - 35%) of N-acetyl-D-glucosamine (GlcNAc) [1, 2]. Among the four types of links occurring between these two kinds of subunits in chitosan, all the enzymes examined for their cleavage specificity acted upon the GlcN-GlcN links. In addition, the chitosanase from Bacillus circulans MH-K1 recognized also GlcN-GlcNAc links [3], while the chitosanase from Streptomyces sp. N174 recognized the GlcNAc-GlcN links [4].
Kinetics and Mechanism
Family GH46 enzymes utilize an inverting mechanism, as shown by NMR [4].
Catalytic Residues
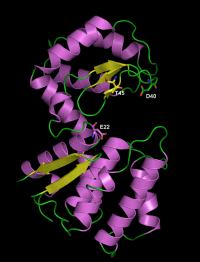
The catalytic residues have been identified by site-directed mutagenesis and crystallography in the chitosanase from Streptomyces sp. N174. The general acid residue is Glu22 in the sequence SSAENSS, while Asp40 (DIGDGRG) is the general base residue [5, 6]. The latter could activate the nucleophilic water molecule with assistance from residue Thr45 (RGYTGGI) [7]. Analysis of sequence alignments as well as crystallographic evidence showed that the same function is played by residues Glu37 (in the sequence NKPEQDD) , Asp55 (DIEDERG) and Thr60 (RGYTIGL) in the chitosanase from Bacillus circulans MH-K1 [8].
Three-dimensional structures
Two structures have been solved using X-ray crystallography, for the chitosanases from Streptomyces sp. N174 [6] and from Bacillus circulans MH-K1 (wild-type enzyme [8] and mutant K218P [9]. These enzymes have essentially an α-helical fold, with two globular domains separated by the active site cleft for substrate binding. The cleft is bordered on the upper face by a three-stranded β-sheet. The structure of GH46 enzymes is similar to the 3D fold of the well studied lysozyme of bacteriophage T4 of Escherichia coli belonging to family GH24 [6] and, to some extent, to the structures of lysozymes from families GH22, GH23 as well the chitinases from family GH19 [10]. These five families are sometimes grouped in the "lysozyme superfamily" [7, 11]. The crystal structures, completed by site-directed mutagenesis have also revealed several residues involved in substrate binding [6, 9, 12, 13]. For the chitosanase from Streptomyces sp N174, the mode of binding of a GlcN hexasaccharide was established as being in conformity with a symmetrical 3+3 model, based on the analysis of products of hydrolysis [12].
Family Firsts
- First primary sequence determination
- Chitosanase from Bacillus circulans MH-K1 [14].
- First sterochemistry determination
- Chitosanase from Streptomyces sp. N174 by NMR [4].
- First general base residue identification
- Chitosanase from Streptomyces sp. N174 by sequence conservation and mutagenesis [5] and by X-ray crystallography [6].
- First general acid residue identification
- Chitosanase from Streptomyces sp. N174 by sequence conservation and mutagenesis [5] and by X-ray crystallography [6].
- First 3-D structure
- Chitosanase from Streptomyces sp. N174 by X-ray crystallography [6].
References
-
Yabuki, M., Uchiyama, A., Suzuki, K., Ando, A., Fujii, T. (1988) Purification and properties of chitosanase from Bacillus circulans MH-K1. Journal of General and Applied Microbiology 34:255-270.
-
Boucher, I., Dupuy, A., Vidal, P., Neugebauer, WA., Brzezinski, R. (1992) Purification and characterization of a chitosanase from Streptomyces N174. Applied Microbiology and Biotechnology 38:188-193.
-
Mitsutomi, M., Ueda, M., Arai, M., Ando, A., Watanabe, T. (1996) Action patterns of microbial chitinases and chitosanases on partially N-acetylated chitosan. Chitin Enzymology, vol. 2, pp 273-284.
- Fukamizo T, Honda Y, Goto S, Boucher I, and Brzezinski R. (1995). Reaction mechanism of chitosanase from Streptomyces sp. N174. Biochem J. 1995;311 ( Pt 2)(Pt 2):377-83. DOI:10.1042/bj3110377 |
- Boucher I, Fukamizo T, Honda Y, Willick GE, Neugebauer WA, and Brzezinski R. (1995). Site-directed mutagenesis of evolutionary conserved carboxylic amino acids in the chitosanase from Streptomyces sp. N174 reveals two residues essential for catalysis. J Biol Chem. 1995;270(52):31077-82. DOI:10.1074/jbc.270.52.31077 |
- Marcotte EM, Monzingo AF, Ernst SR, Brzezinski R, and Robertus JD. (1996). X-ray structure of an anti-fungal chitosanase from streptomyces N174. Nat Struct Biol. 1996;3(2):155-62. DOI:10.1038/nsb0296-155 |
- Lacombe-Harvey ME, Fukamizo T, Gagnon J, Ghinet MG, Dennhart N, Letzel T, and Brzezinski R. (2009). Accessory active site residues of Streptomyces sp. N174 chitosanase: variations on a common theme in the lysozyme superfamily. FEBS J. 2009;276(3):857-69. DOI:10.1111/j.1742-4658.2008.06830.x |
- Saito J, Kita A, Higuchi Y, Nagata Y, Ando A, and Miki K. (1999). Crystal structure of chitosanase from Bacillus circulans MH-K1 at 1.6-A resolution and its substrate recognition mechanism. J Biol Chem. 1999;274(43):30818-25. DOI:10.1074/jbc.274.43.30818 |
- Fukamizo T, Amano S, Yamaguchi K, Yoshikawa T, Katsumi T, Saito J, Suzuki M, Miki K, Nagata Y, and Ando A. (2005). Bacillus circulans MH-K1 chitosanase: amino acid residues responsible for substrate binding. J Biochem. 2005;138(5):563-9. DOI:10.1093/jb/mvi156 |
- Monzingo AF, Marcotte EM, Hart PJ, and Robertus JD. (1996). Chitinases, chitosanases, and lysozymes can be divided into procaryotic and eucaryotic families sharing a conserved core. Nat Struct Biol. 1996;3(2):133-40. DOI:10.1038/nsb0296-133 |
- Holm L and Sander C. (1994). Structural similarity of plant chitinase and lysozymes from animals and phage. An evolutionary connection. FEBS Lett. 1994;340(1-2):129-32. DOI:10.1016/0014-5793(94)80187-8 |
- Tremblay H, Yamaguchi T, Fukamizo T, and Brzezinski R. (2001). Mechanism of chitosanase-oligosaccharide interaction: subsite structure of Streptomyces sp. N174 chitosanase and the role of Asp57 carboxylate. J Biochem. 2001;130(5):679-86. DOI:10.1093/oxfordjournals.jbchem.a003034 |
- Katsumi T, Lacombe-Harvey ME, Tremblay H, Brzezinski R, and Fukamizo T. (2005). Role of acidic amino acid residues in chitooligosaccharide-binding to Streptomyces sp. N174 chitosanase. Biochem Biophys Res Commun. 2005;338(4):1839-44. DOI:10.1016/j.bbrc.2005.10.157 |
-
Ando, A., Noguchi, K., Yanagi, M., Shinoyama, H., Kagawa, Y., Hirata, H., Yabuki, M., Fujii, T. (1992) Primary structure of chitosanase produced by Bacillus circulans MH-K1. Journal of General and Applied Microbiology 38:135-144.