CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Glycoside Hydrolase Family 117"
| Line 52: | Line 52: | ||
;First catalytic nucleophile identification: - | ;First catalytic nucleophile identification: - | ||
;First general acid/base residue identification: - | ;First general acid/base residue identification: - | ||
| − | ;First 3-D structure: Zg4663, α-1,3-L-(3,6-anhydro)-galactosidase (AhgA), PDB: [http://www.pdb.org/pdb/explore/explore.do?structureId=3p2n 3P2N] <cite>Rebuffet2011</cite>. | + | ;First 3-D structure: 2011: Zg4663, α-1,3-L-(3,6-anhydro)-galactosidase (AhgA), PDB: [http://www.pdb.org/pdb/explore/explore.do?structureId=3p2n 3P2N] <cite>Rebuffet2011</cite>. |
Revision as of 06:34, 5 May 2011
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^Etienne Rebuffet^^^
- Responsible Curator: ^^^Mirjam Czjzek^^^
| Glycoside Hydrolase Family GH117 | |
| Clan | None |
| Mechanism | Not known |
| Active site residues | Not known |
| CAZy DB link | |
| http://www.cazy.org/GH117.html | |
Substrate specificities
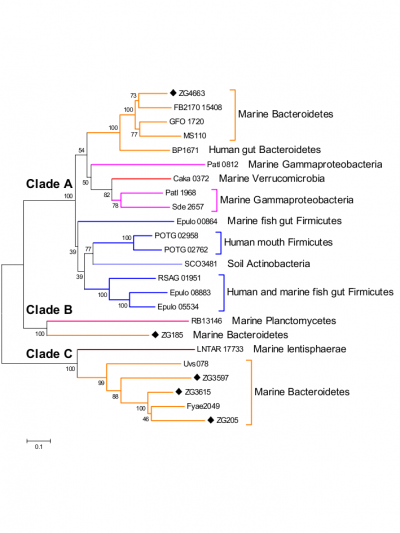
The only activity so far identified in this recently discovered family of glycoside hydrolases is that α-1,3-L-(3,6-anhydro)-galactosidase [1, 2, 3]. Nevertheless phylogenetic analysis (figure 1) of this family and activity test on Zg3597 (Clade C) show that the family GH117 is polyspecific [1].
Kinetics and Mechanism
Mechanism of glycoside hydrolase family 117 is still unknown. But structural analyse revealed the presence of a zinc ion, only coordinated by water molecules, close to the active site, which could activate the catalytic water molecule and provide the energy needed for the enzymatic reaction to take place [1]. Sequence alignment suggest that the enzymes of clades B and C do not bind zinc ions which could be related to the difference of substrate.
Catalytic Residues
From structural analysis and sequences alignment the catalytic residues have been predicted to be two out of the three acidic residues Asp-97, Asp-252 and Glu-310 (Zg4663 numbering) [1].
Three-dimensional structures
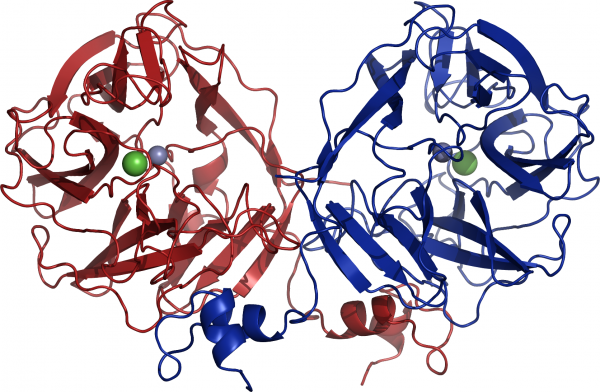
At the moment two members of GH117 family have been crystallized. Both are enzymes from marine bacteria, one from Saccharophagus degradans [4] and one from Zobelia galactanivorans [1]. A crystal structure has only been reported for the α-1,3-L-(3,6-anhydro)-galactosidase (AhgA, Zg4663) from Z. galactanivorans (PDB: 3P2N) [1]. Zg4663 adopting the five-bladed β-propeller fold and form dimer via domain-swapping of the N-terminal HTH (Helix-Turn-Helix) domain (Figure 2) [1]. Interestingly, previous sequences reported from Vibrio sp. JT0107 and Bacillus sp. MK03 contain the conserved domain-swapping signature SxAxxR in the HTH domain. Consistently, these proteins were reported to form multimers (a dimer and an octamer respectively), based on calibrated gel filtration estimations [2, 3]. In contrast, RB13146 (Clade B) misses the domain-swapping signature, and the crucial residues are missing. This protein from R. baltica is thus likely a monomer and may represent an ‘ancestral’ form of the GH117 family which would be limited to the β-propeller, catalytic domain [1].
Family Firsts
- First stereochemistry determination
- -
- First catalytic nucleophile identification
- -
- First general acid/base residue identification
- -
- First 3-D structure
- 2011: Zg4663, α-1,3-L-(3,6-anhydro)-galactosidase (AhgA), PDB: 3P2N [1].
References
- Rebuffet E, Groisillier A, Thompson A, Jeudy A, Barbeyron T, Czjzek M, and Michel G. (2011). Discovery and structural characterization of a novel glycosidase family of marine origin. Environ Microbiol. 2011;13(5):1253-70. DOI:10.1111/j.1462-2920.2011.02426.x |
- Sugano Y, Kodama H, Terada I, Yamazaki Y, and Noma M. (1994). Purification and characterization of a novel enzyme, alpha-neoagarooligosaccharide hydrolase (alpha-NAOS hydrolase), from a marine bacterium, Vibrio sp. strain JT0107. J Bacteriol. 1994;176(22):6812-8. DOI:10.1128/jb.176.22.6812-6818.1994 |
- Suzuki H, Sawai Y, Suzuki T, and Kawai K. (2002). Purification and characterization of an extracellular alpha-neoagarooligosaccharide hydrolase from Bacillus sp. MK03. J Biosci Bioeng. 2002;93(5):456-63. DOI:10.1016/s1389-1723(02)80092-5 |
- Lee S, Lee JY, Ha SC, Jung J, Shin DH, Kim KH, and Choi IG. (2009). Crystallization and preliminary X-ray analysis of neoagarobiose hydrolase from Saccharophagus degradans 2-40. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65(Pt 12):1299-301. DOI:10.1107/S174430910904603X |