CAZypedia celebrates the life of Senior Curator Emeritus Harry Gilbert, a true giant in the field, who passed away in September 2025.
CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article. Totally new to the CAZy classification? Read this first.
Glycoside Hydrolase Family 18
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
| Glycoside Hydrolase Family GH18 | |
| Clan | GH-K |
| Mechanism | retaining |
| Active site residues | known (acid/neighbouring group) |
| CAZy DB link | |
| https://www.cazy.org/GH18.html | |
Substrate specificities
Family GH18 is unusual in having glycoside hydrolases that are both catalytically active chitinases (EC 3.2.1.14) and endo-β-N-acetylglucosaminidases (EC 3.2.1.96) and also sub-families of non-hydrolytic proteins that function as carbohydrate binding modules / "lectins" or as xylanase inhibitors.
The active chitinases comprise non-processive endo-acting enzymes as well as processive enzymes with exo- and endo-binding preferences [1, 2]. Most enzymes primarily produce chitobiose, but some endo-acting family 18 chitinases are not capable of cleaving trimers or tetramers and thus yield longer products. Note that in older literature the endo-/exo-character of these enzymes often is assessed by studying the degradation of oligomeric substrates, and that several recent studies have shown this method to be invalid.
Family 18 chitinases break down all forms of chitin at varying rates depending on the enzyme and the substrate. They also act on chitosan with degrees of acetylation as low as 13 % [3] and some are known to degrade peptidoglycan [4]. Studies on family 18 chitinases from Serratia marcescens have suggested that most subsites for sugar binding show some promiscuity with the notable exception of subsite -1 [1].
Kinetics and Mechanism
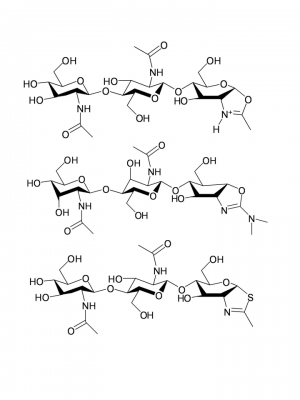
GH18 enzymes perform enzymic catalysis with retention of anomeric configuration. They belong to a group of enzymes (including GH families 18, 20, 25, 56, 84, and 85) that perform catalysis using a double-displacement reaction but instead of the more common enzyme-derived nucleophile they utilize the N-acetamido carbonyl oxygen in what is termed neighboring group participation (or substrate participation or anchimeric assistance). Figures showing such a mechanism date back to Koshland's 1953 review [5]; indeed they frequent the chemical literature of participating groups long before that, but it is primarily through the work on GH18 [6] and soon after GH20 [7, 8] that such a mechanism became well established. In such a mechanism, which occurs with (net) retention of anomeric configuration, the enzyme provides a general acid function to protonate the leaving group to facilitate its departure with the substrate carbonyl oxygen playing the role of nucleophile to generate a bicyclic "oxazolinium ion" intermediate, which subsequently breaks down following the microscopic reverse via hydrolysis or occasionally transglycosylation). Such a mechanism has a number of facets, one of which is its potential inhibition using thiazolines [9]. Allosamidin, a pseudotrisaccharide consisting of two N-acetylallosamine sugars linked to an allosamizoline moiety [10], is a well known natural compound that is a high affinity inhibitor of family 18 chitinases (see Figure).
Like in many other enzymes acting on polysaccharides the substrate-binding clefts of processive family 18 chitinases are lined with aromatic residues. Family 18 chitinases have proven very useful to gain insight into the structural basis of a processive mechanism and in the importance of such a mechanism for biomass-converting efficiency [11, 12]. As previously suggested for cellulases in e.g. [13], aromatic residues close to the catalytic center play a crucial role in processivity [14].
Catalytic Residues
The catalytically-active GH18 enzymes use a double displacement reaction mechanism with neighboring group participation(see Figure). In this mechanism the carbonyl oxygen of the substrate acts as a nucleophile, with assistance from a carboxylate (Asp) that acts to deprotonate the N-acetamido nitrogen during oxazolinium ion formation/breakdown. A second catalytic acid residue (glutamate in family GH18, but often also Asp in other families using this mechanism) acts as a general acid/base to protonate the glycosidic oxygen to assist in the departure of the aglycon, and to deprotonate the nucleophilic water molecule during the hydrolysis of the oxazolinium ion intermediate. In family GH18 the two catalytic carboxylates are found in an D-X-E motif whereas in other families the carboxylates may be adjacent, such as the DD motif in family GH84 (for example see [15]). The physical separation of the two catalytic residues (with the second not in a position to act as a nucleophile itself) has led to confusion in some literature that GH18 and other enzymes (notably GH25) may be inverting enzymes; this is certainly not the case for GH18 and is unlikely to be the case for GH25.
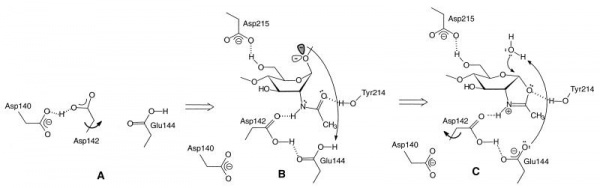
The D-X-E motif (Asp140-Glu144 in the Figure) is part of a diagnostic D-X-X-D-X-D-X-E motif that includes two more aspartates of which the one in italics (Asp140 in the Figure) is known to be essential for catalytic activity. There are several other conserved residues in the catalytic center that play important roles during catalysis, related to distortion of the -1 sugar, activation of the acetamido group and/or cycling of the pKa of the catalytic glutamate [17]. The O6 of the –1 sugar interacts with the side chain of yet another semi-conserved aspartate (Asp215 in the Figure). In enzymes with an acidic pH optimum this residue is an asparagine.
Catalytically inactive members
One unusual of feature of plant members of the GH18 family is the large number of sequences that encode catalytically-inactive proteins that function as enzyme inhibitors or lectins. Phylogenetic analysis of the plant GH18 family reveals clear distinction between hevamine-type chitinases, putative chitinases and narbonins [18]. Out of the major subfamilies, only the one that contains hevamine contains enzymes of demonstrated activity [19]. The subfamily of GH18 that contains xylanase inhibitor proteins (XIP) emerged from the hevamine cluster along with concanavalin B. All have non-conservative substitutions of one of the acidic amino acid residues in the catalytic region. In the structure of concanavalin B the catalytic Glu residue is replaced by Gln [20], which mostly accounts for the lack of chitinase activity reported for this protein. The XIP-type inhibitors all have the third aspartic acid DxxDxDxE mutated into an aromatic residue whereas the catalytic glutamate residue is only conserved in the prototype of cereal xylanase inhibitors, XIP-I (isolated from Triticum aetivum) [21]. In XIP-I and narbonin, the glutamic acid residues are present in an equivalent position to the catalytic residue in hevamine, but their side chain is fully engaged in salt bridges with neighbouring arginine residues [22, 23], preventing chitinase activity despite the presence of the catalytic residue. Furthermore in both XIP-I and narbonin, the position equivalent to residue Asp in hevamine, which has been proposed to stabilize the positively charged oxazolinium ion reaction intermediate [19], is occupied by a bulky residue [22, 23]. The mutation of this Asp residue in alanine in hevamine led to a mutant with approx. 2% residual activity [24]. The most striking disruption of the cleft in XIP-I and narbonin is caused by the mutation of subsite -1 Gly which participates in the hydrogen-bonding network with the ligand [6, 25], resulting in complete obstruction of subsite -1 and preventing access to the catalytic residue [23]. Xylanase inhibitors appeared after the emergence of the various subfamilies of chitinases from their common ancestor. In this respect, the xylanase inhibitors are a relatively new acquisition, and so far no protein has been reported to display both xylanase inhibition and chitinase activities. GH18 XIP-type inhibitors can inhibit xylanases from GH10 and GH11 families [21]. The inhibition specificity of the GH18 xylanase inhibitors can be explained on the basis of the solved 3-D structure of XIP-I in complex with a GH10 xylanase from A. nidulans and a GH11 xylanase from P. funiculosum [26].
Three-dimensional structures
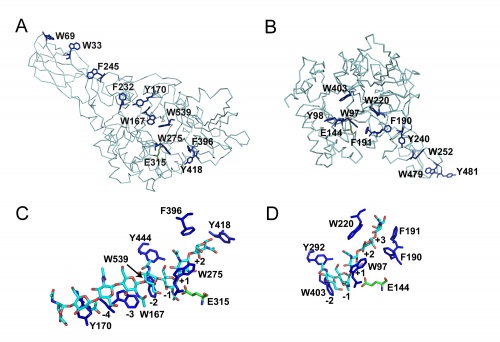
Although these enzymes are frequently multi-modular, the catalytic domains are α / β barrels [25, 27]. While several structures for complete bi-modular chitinases are available [27, 28] (see Figure), available structural information for multi-modular enzymes is often limited to the isolated catalytic domain. Work on the conformational itinerary of catalysis which is extremely similar to other retaining enzymes active on gluco-configured substrates, was provided by the van Aalten group [16] in 2001 through the trapping of a distorted Michaelis complex in 1,4B conformation and thus extremely similar to the 1S3 skew boats observed in GH5 [29] for example or the 4E conformation originally seen for a "neighboring group" enzyme in GH20 [7]. More recently, a similar conformation has been observed for the Michaelis complex of another neighboring group enzyme, the GH84 O-GlcNAcase [15]. Fungal GH18 enzymes are considered as possible therapeutic targets and a number of programmes are probing this area (for example [30]).
Family Firsts
- First sterochemistry determination
- Sometimes incorrectly reported as inverting, this family performs catalysis with retention of anomeric configuration as first shown on the Bacillus ciculans enzyme [31].
- First catalytic nucleophile identification
- This family is one of many that uses neighbouring group participation for catalysis with the N-acetyl carbonyl group acting as the nucleophile; first proposed (we believe) for this family in [6].
- First general acid/base residue identification
- On the basis of 3-D structure [27].
- First 3-D structure
- The first two 3-D structures for catalytically active GH18 members were the Serratia marcescens chitinase A and the plant defence protein hevamine published "back-to-back" in Structure in 1994 [25, 27]. In retrospect, however, the non-catalytic "narbonin" structure was arguably the first GH18 3-D structure, although it is has no enzymatic activity [32].
References
- Horn SJ, Sørbotten A, Synstad B, Sikorski P, Sørlie M, Vårum KM, and Eijsink VG. (2006). Endo/exo mechanism and processivity of family 18 chitinases produced by Serratia marcescens. FEBS J. 2006;273(3):491-503. DOI:10.1111/j.1742-4658.2005.05079.x |
- Hult EL, Katouno F, Uchiyama T, Watanabe T, and Sugiyama J. (2005). Molecular directionality in crystalline beta-chitin: hydrolysis by chitinases A and B from Serratia marcescens 2170. Biochem J. 2005;388(Pt 3):851-6. DOI:10.1042/BJ20050090 |
- Sørbotten A, Horn SJ, Eijsink VG, and Vårum KM. (2005). Degradation of chitosans with chitinase B from Serratia marcescens. Production of chito-oligosaccharides and insight into enzyme processivity. FEBS J. 2005;272(2):538-49. DOI:10.1111/j.1742-4658.2004.04495.x |
- Bokma E, van Koningsveld GA, Jeronimus-Stratingh M, and Beintema JJ. (1997). Hevamine, a chitinase from the rubber tree Hevea brasiliensis, cleaves peptidoglycan between the C-1 of N-acetylglucosamine and C-4 of N-acetylmuramic acid and therefore is not a lysozyme. FEBS Lett. 1997;411(2-3):161-3. DOI:10.1016/s0014-5793(97)00682-0 |
-
Koshland, D. (1953) Biol. Rev. 28, 416.
- Terwisscha van Scheltinga AC, Armand S, Kalk KH, Isogai A, Henrissat B, and Dijkstra BW. (1995). Stereochemistry of chitin hydrolysis by a plant chitinase/lysozyme and X-ray structure of a complex with allosamidin: evidence for substrate assisted catalysis. Biochemistry. 1995;34(48):15619-23. DOI:10.1021/bi00048a003 |
- Tews I, Perrakis A, Oppenheim A, Dauter Z, Wilson KS, and Vorgias CE. (1996). Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nat Struct Biol. 1996;3(7):638-48. DOI:10.1038/nsb0796-638 |
- Drouillard S, Armand S, Davies GJ, Vorgias CE, and Henrissat B. (1997). Serratia marcescens chitobiase is a retaining glycosidase utilizing substrate acetamido group participation. Biochem J. 1997;328 ( Pt 3)(Pt 3):945-9. DOI:10.1042/bj3280945 |
- Macdonald JM, Tarling CA, Taylor EJ, Dennis RJ, Myers DS, Knapp S, Davies GJ, and Withers SG. (2010). Chitinase inhibition by chitobiose and chitotriose thiazolines. Angew Chem Int Ed Engl. 2010;49(14):2599-602. DOI:10.1002/anie.200906644 |
- Sakuda S, Isogai A, Matsumoto S, and Suzuki A. (1987). Search for microbial insect growth regulators. II. Allosamidin, a novel insect chitinase inhibitor. J Antibiot (Tokyo). 1987;40(3):296-300. DOI:10.7164/antibiotics.40.296 |
- Eijsink VG, Vaaje-Kolstad G, Vårum KM, and Horn SJ. (2008). Towards new enzymes for biofuels: lessons from chitinase research. Trends Biotechnol. 2008;26(5):228-35. DOI:10.1016/j.tibtech.2008.02.004 |
- Zakariassen H, Aam BB, Horn SJ, Vårum KM, Sørlie M, and Eijsink VG. (2009). Aromatic residues in the catalytic center of chitinase A from Serratia marcescens affect processivity, enzyme activity, and biomass converting efficiency. J Biol Chem. 2009;284(16):10610-7. DOI:10.1074/jbc.M900092200 |
- Varrot A, Frandsen TP, von Ossowski I, Boyer V, Cottaz S, Driguez H, Schülein M, and Davies GJ. (2003). Structural basis for ligand binding and processivity in cellobiohydrolase Cel6A from Humicola insolens. Structure. 2003;11(7):855-64. DOI:10.1016/s0969-2126(03)00124-2 |
- Horn SJ, Sikorski P, Cederkvist JB, Vaaje-Kolstad G, Sørlie M, Synstad B, Vriend G, Vårum KM, and Eijsink VG. (2006). Costs and benefits of processivity in enzymatic degradation of recalcitrant polysaccharides. Proc Natl Acad Sci U S A. 2006;103(48):18089-94. DOI:10.1073/pnas.0608909103 |
- He Y, Macauley MS, Stubbs KA, Vocadlo DJ, and Davies GJ. (2010). Visualizing the reaction coordinate of an O-GlcNAc hydrolase. J Am Chem Soc. 2010;132(6):1807-9. DOI:10.1021/ja9086769 |
- van Aalten DM, Komander D, Synstad B, Gåseidnes S, Peter MG, and Eijsink VG. (2001). Structural insights into the catalytic mechanism of a family 18 exo-chitinase. Proc Natl Acad Sci U S A. 2001;98(16):8979-84. DOI:10.1073/pnas.151103798 |
- Synstad B, Gåseidnes S, Van Aalten DM, Vriend G, Nielsen JE, and Eijsink VG. (2004). Mutational and computational analysis of the role of conserved residues in the active site of a family 18 chitinase. Eur J Biochem. 2004;271(2):253-62. DOI:10.1046/j.1432-1033.2003.03923.x |
- Durand A, Hughes R, Roussel A, Flatman R, Henrissat B, and Juge N. (2005). Emergence of a subfamily of xylanase inhibitors within glycoside hydrolase family 18. FEBS J. 2005;272(7):1745-55. DOI:10.1111/j.1742-4658.2005.04606.x |
- Terwisscha van Scheltinga AC, Hennig M, and Dijkstra BW. (1996). The 1.8 A resolution structure of hevamine, a plant chitinase/lysozyme, and analysis of the conserved sequence and structure motifs of glycosyl hydrolase family 18. J Mol Biol. 1996;262(2):243-57. DOI:10.1006/jmbi.1996.0510 |
- Hennig M, Jansonius JN, Terwisscha van Scheltinga AC, Dijkstra BW, and Schlesier B. (1995). Crystal structure of concanavalin B at 1.65 A resolution. An "inactivated" chitinase from seeds of Canavalia ensiformis. J Mol Biol. 1995;254(2):237-46. DOI:10.1006/jmbi.1995.0614 |
- Juge N, Payan F, and Williamson G. (2004). XIP-I, a xylanase inhibitor protein from wheat: a novel protein function. Biochim Biophys Acta. 2004;1696(2):203-11. DOI:10.1016/j.bbapap.2003.08.014 |
- Hennig M, Pfeffer-Hennig S, Dauter Z, Wilson KS, Schlesier B, and Nong VH. (1995). Crystal structure of narbonin at 1.8 A resolution. Acta Crystallogr D Biol Crystallogr. 1995;51(Pt 2):177-89. DOI:10.1107/S0907444994009807 |
- Payan F, Flatman R, Porciero S, Williamson G, Juge N, and Roussel A. (2003). Structural analysis of xylanase inhibitor protein I (XIP-I), a proteinaceous xylanase inhibitor from wheat (Triticum aestivum, var. Soisson). Biochem J. 2003;372(Pt 2):399-405. DOI:10.1042/BJ20021802 |
- Bokma E, Rozeboom HJ, Sibbald M, Dijkstra BW, and Beintema JJ. (2002). Expression and characterization of active site mutants of hevamine, a chitinase from the rubber tree Hevea brasiliensis. Eur J Biochem. 2002;269(3):893-901. DOI:10.1046/j.0014-2956.2001.02721.x |
- Terwisscha van Scheltinga AC, Kalk KH, Beintema JJ, and Dijkstra BW. (1994). Crystal structures of hevamine, a plant defence protein with chitinase and lysozyme activity, and its complex with an inhibitor. Structure. 1994;2(12):1181-9. DOI:10.1016/s0969-2126(94)00120-0 |
- Payan F, Leone P, Porciero S, Furniss C, Tahir T, Williamson G, Durand A, Manzanares P, Gilbert HJ, Juge N, and Roussel A. (2004). The dual nature of the wheat xylanase protein inhibitor XIP-I: structural basis for the inhibition of family 10 and family 11 xylanases. J Biol Chem. 2004;279(34):36029-37. DOI:10.1074/jbc.M404225200 |
- Perrakis A, Tews I, Dauter Z, Oppenheim AB, Chet I, Wilson KS, and Vorgias CE. (1994). Crystal structure of a bacterial chitinase at 2.3 A resolution. Structure. 1994;2(12):1169-80. DOI:10.1016/s0969-2126(94)00119-7 |
- van Aalten DM, Amadei A, Linssen AB, Eijsink VG, Vriend G, and Berendsen HJ. (1995). The essential dynamics of thermolysin: confirmation of the hinge-bending motion and comparison of simulations in vacuum and water. Proteins. 1995;22(1):45-54. DOI:10.1002/prot.340220107 |
- Davies GJ, Mackenzie L, Varrot A, Dauter M, Brzozowski AM, Schülein M, and Withers SG. (1998). Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase. Biochemistry. 1998;37(34):11707-13. DOI:10.1021/bi981315i |
- Houston DR, Shiomi K, Arai N, Omura S, Peter MG, Turberg A, Synstad B, Eijsink VG, and van Aalten DM. (2002). High-resolution structures of a chitinase complexed with natural product cyclopentapeptide inhibitors: mimicry of carbohydrate substrate. Proc Natl Acad Sci U S A. 2002;99(14):9127-32. DOI:10.1073/pnas.132060599 |
- Armand S, Tomita H, Heyraud A, Gey C, Watanabe T, and Henrissat B. (1994). Stereochemical course of the hydrolysis reaction catalyzed by chitinases A1 and D from Bacillus circulans WL-12. FEBS Lett. 1994;343(2):177-80. DOI:10.1016/0014-5793(94)80314-5 |
- Hennig M, Schlesier B, Dauter Z, Pfeffer S, Betzel C, Höhne WE, and Wilson KS. (1992). A TIM barrel protein without enzymatic activity? Crystal-structure of narbonin at 1.8 A resolution. FEBS Lett. 1992;306(1):80-4. DOI:10.1016/0014-5793(92)80842-5 |
- Houston DR, Recklies AD, Krupa JC, and van Aalten DM. (2003). Structure and ligand-induced conformational change of the 39-kDa glycoprotein from human articular chondrocytes. J Biol Chem. 2003;278(32):30206-12. DOI:10.1074/jbc.M303371200 |