CAZypedia needs your help!
We have many unassigned pages in need of Authors and Responsible Curators. See a page that's out-of-date and just needs a touch-up? - You are also welcome to become a CAZypedian. Here's how.
Scientists at all career stages, including students, are welcome to contribute.
Learn more about CAZypedia's misson here and in this article.
Totally new to the CAZy classification? Read this first.
Difference between revisions of "Syn/anti lateral protonation"
| Line 1: | Line 1: | ||
| − | |||
| − | |||
* [[Author]]: ^^^Wim Nerinckx^^^ | * [[Author]]: ^^^Wim Nerinckx^^^ | ||
* [[Responsible Curator]]: ^^^Spencer Williams^^^ | * [[Responsible Curator]]: ^^^Spencer Williams^^^ | ||
Revision as of 20:29, 14 November 2012
- Author: ^^^Wim Nerinckx^^^
- Responsible Curator: ^^^Spencer Williams^^^
Overview
This page provides a table that summarizes the spatial positioning of the catalytic general acid residue in the active sites of glycoside hydrolases, relative to the substrate. The table below updates those found in the seminal paper on this concept by Heightman and Vasella [1], and a following paper by Nerinckx et al. [2].
Background
The "not from above, but from the side" concept of semi-lateral glycosidic oxygen protonation by glycoside hydrolases was introduced by Heightman and Vasella [1]. It was originally only described for beta-equatorial glycoside hydrolases, but appears to be equally applicable to enzymes acting on an alpha-axial glycosidic bond [2]. When dividing subsite -1 into half-spaces by a plane defined by the glycosidic oxygen and C1' and H1' of the –1 glycoside, many ligand-complexed structures reveal that the proton donor is positioned either in the syn half-space (near the ring-oxygen of the –1 glycoside), or in the anti half-space (on the opposite side of the ring-oxygen). Members of the same GH family appear to share a common syn or anti protonator arrangement and further, this specificity appears to be preserved within Clans of families. This page's compilation of subsite -1 occupied complexes shows that about 70% of all GH families are anti protonators.
Closer inspection of –1/+1 subsite-spanning, substrate or substrate-analogue ligands in crystal structures reveals a further intriguing corollary [2], [3]. In substrate-bound complexes with anti protonating GH enzymes, the scissile anomeric bond (often studied using the thio-analogue) show a dihedral angle φ (O5'-C1'-[O,S]x-Cx) that is in the lowest-energy synclinal (gauche) arrangement. A rationale for this is that a minus synclinal dihedral angle φ for an equatorial glycosidic bond, or plus synclinal for an axial bond [4], allows for hyperconjugative overlap of the C1'-O5' antibonding orbital with an antiperiplanar-oriented lone pair orbital lobe of the glycosidic oxygen, thereby creating partial double bond character and stabilization of the glycosidic bond by 4–5 kcal/mol; this ground-state stabilizing phenomenon is known as the ‘exo-anomeric effect’ [5] [6]. Anti protonation results in interaction with the glycosidic oxygen’s antiperiplanar lone pair, which removes the stabilizing anomeric effect. This suggests that anti-protonation is an enzymic approach for lowering the activation barrier leading to the transition state (Figure 1 centre).
Syn protonating GHs show a rather different arrangement of the anomeric substituent of substrates or substrate-analogues [2], [3]. In many –1/+1 subsite-spanning ligand complexes the dihedral angle φ of the scissile bond has been rotated away from its lowest-energy synclinal position: clockwise to minus-anticlinal or antiperiplanar for beta-equatorial; counterclockwise to plus-anticlinal or antiperiplanar for alpha-axial anomeric bonds. This removes the hyperconjugative overlap resulting in the stabilizing exo-anomeric effect. Because of this rotation, the lone pair of the glycosidic oxygen points into the syn half-space, and is that protonated by the syn-positioned proton donor (Figure 1 right).
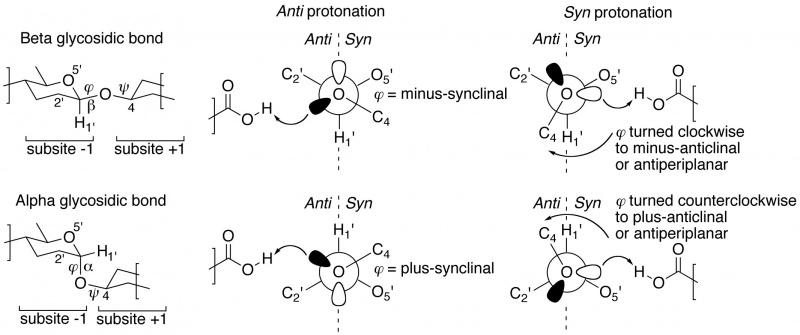
Table of syn/anti protonation examples
Note
This table contains only one example per GH family of a ligand-complexed protein structure where the syn positioning or anti positioning of the proton donor can be clearly observed; other examples may be available on a family-by-family basis. The reader is thus advised to consult the CAZy database for a current, comprehensive list of CAZyme structures. Where available, the selected examples are Michaelis-type complexes with the ligand spanning the -1/+1 subsites, since these have an intact glycosidic or thioglycosidic bond, or are N-analogs of the substrate (e.g. acarbose). In some examples, the proton donor has been mutated (e.g., to the corresponding amide or to an alanine), and in those cases one may wish to look at a superposition of the given PDB example with the structure of the native enzyme. If a Michaelis-type complex is not yet available, the second and third example choices, respectively, are trapped glycosyl-enzyme intermediates and product complexes where subsite -1 is occupied.
Please also be aware that this is a large table with many data. Please contact the page Author or Responsible Curator with corrections.
Table
This table can be re-sorted by clicking on the icons in the header (javascript must be turned on in your browser). To reset the page to be sorted by GH family, click the page tab at the very top of the page.
| Family | Clan | Structure fold | Anomeric specificity | Mechanism | Syn/anti protonator | Example PDB ID | Enzyme | Organism | Ligand | General acid | Nucleophile or General base | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GH1 | A | (β/α)8 | beta | retaining | anti | 2cer | β-glycosidase S | Sulfolobus solfataricus P2 | phenethyl glucoimidazol | Glu206 | Glu387 | [7] |
| GH2 | A | (β/α)8 | beta | retaining | anti | 2vzu | exo-β-glucosaminidase | Amicolatopsis orientalis | PNP-β-d-glucosamine | Glu469 | Glu541 | [8] |
| GH3 | none | (β/α)8 | beta | retaining | anti | 1iex | exo-1,3-1,4-glucanase | Hordeum vulgare | thiocellobiose | Glu491 | Asp285 | [9] |
| GH5 | A | (β/α)8 | beta | retaining | anti | 1h2j | endo-β-1,4-glucanase | Bacillus agaradhaerens | 2',4'-DNP-2-F-cellobioside | Glu129 | Glu228 | [10] |
| GH6 | none | (β/α)8 | beta | inverting | syn | 1qjw | cellobiohydrolase 2 | Hypocrea jecorina | (Glc)2-S-(Glc)2 | Asp221 | debated | [11] |
| GH7 | B | β-jelly roll | beta | retaining | syn | 1ovw | endo-1,4-glucanase | Fusarium oxysporum | thio-(Glc)5 | Glu202 | Glu197 | [12] |
| GH8 | M | (α/α)6 | beta | inverting | anti | 1kwf | endo-1,4-glucanase | Clostridium thermocellum | cellopentaose | Glu95 | Asp278 | [13] |
| GH9 | none | (α/α)6 | beta | inverting | syn | 1rq5 | cellobiohydrolase | Clostridium thermocellum | cellotetraose | Glu795 | Asp383 | [14] |
| GH10 | A | (β/α)8 | beta | retaining | anti | 2d24 | β-1,4-xylanase | Streptomyces olivaceoviridis E-86 | xylopentaose | Glu128 | Glu236 | [15] |
| GH11 | C | β-jelly roll | beta | retaining | syn | 1bvv | xylanase | Bacillus circulans | Xyl-2-F-xylosyl | Glu172 | Glu78 | [16] |
| GH12 | C | β-jelly roll | beta | retaining | syn | 1w2u | endoglucanase | Humicola grisea | thiocellotetraose | Glu205 | Glu120 | [17] |
| GH13 | H | (β/α)8 | alpha | retaining | anti | 1cxk | β-cyclodextrin glucanotransferase | Bacillus circulans | maltononaose | Glu257 | Asp229 | [18] |
| GH14 | none | (β/α)8 | alpha | inverting | syn | 1itc | β-amylase | Bacillus cereus | maltopentaose | Glu172 | Glu367 | [19] |
| GH15 | L | (α/α)6 | alpha | inverting | syn | 1gah | glucoamylase | Aspergillus awamori | acarbose | Glu179 | Glu400 | [20] |
| GH16 | B | β-jelly roll | beta | retaining | syn | 1urx | β-agarase A | Zobellia galactanivorans | oligoagarose | Glu152 | Glu147 | [21] |
| GH17 | A | (β/α)8 | beta | retaining | predicted anti by clan | see e.g. at GH1 | ||||||
| GH18 | K | (β/α)8 | beta | retaining | anti | 1ffr | chitinase A | Serratia marcescens | (NAG)6 | Glu315 | internal | [22] |
| GH20 | K | (β/α)8 | beta | retaining | anti | 1c7s | chitobiase | Serratia marcescens | chitobiose | Glu540 | internal | [23] |
| GH22 | none | lysozyme type | beta | retaining | syn | 1h6m | lysozyme C | Gallus gallus | Chit-2-F-chitosyl | Glu35 | Asp52 | [24] |
| GH23 | none | lysozyme type | beta | inverting | syn | 1lsp | lysozyme G | Cygnus atratus | Bulgecin A | Glu73 | internal | [25] |
| GH24 | I | α + β | beta | inverting | syn | 148l | lysozyme E | Bacteriophage T4 | chitobiosyl | Glu11 | Glu26 | [26] |
| GH26 | A | (β/α)8 | beta | retaining | anti | 1gw1 | mannanase A | Cellvibrio japonicus | 2',4'-DNP-2-F-cellotrioside | Glu212 | Glu320 | [27] |
| GH27 | D | (β/α)8 | alpha | retaining | anti | 1ktc | α-N-acetyl galactosaminidase | Gallus gallus | NAGal | Asp201 | Asp410 | [28] |
| GH28 | N | β-helix | alpha | inverting | anti | 2uvf | exo-polygalacturonosidase | Yersinia enterocolitica ATCC9610D | digalacturonic acid | Asp402 | Asp381 Asp403 | [29] |
| GH29 | none | (β/α)8 | alpha | retaining | syn | 1hl9 | α-l-fucosidase | Thermotoga maritima | 2-F-fuco- pyranosyl | Glu266 | Asp224 | [30] |
| GH30 | A | (β/α)8 | beta | retaining | anti | 2v3d | glucocerebrosidase 1 | Homo sapiens | N-butyl-deoxynojirimycin | Glu235 | Glu340 | [31] |
| GH31 | D | (β/α)8 | alpha | retaining | anti | 2qmj | maltase-glucoamylase | Homo sapiens | acarbose | Asp542 | Asp443 | [32] |
| GH32 | J | 5-fold β-propeller | beta | retaining | anti | 2add | fructan β-(2,1)-fructosidase | Cichorium intybus | sucrose | Glu201 | Asp22 | [33] |
| GH33 | E | 6-fold β-propeller | alpha | retaining | anti | 1s0i | trans-sialidase | Trypanosoma cruzi | sialyl-lactose | Asp59 | Tyr342 | [34] |
| GH34 | E | 6-fold β-propeller | alpha | retaining | anti | 2bat | neuraminidase | Influenza A virus | sialic acid | Asp151 | Tyr406 | [35] |
| GH35 | A | (β/α)8 | beta | retaining | anti | 1xc6 | β-galactosidase | Penicillium sp. | d-galactose | Glu200 | Glu299 | [36] |
| GH37 | G | (α/α)6 | alpha | inverting | anti | 2jf4 | trehalase | Escherechia coli | validoxylamine | Asp312 | Glu496 | [37] |
| GH38 | none | (β/α)7 | alpha | retaining | anti | 1qwn | α-mannosidase II | Drosophila melanogaster | 5-F-β-l-gulosyl | Asp341 | Asp204 | [38] |
| GH39 | A | (β/α)8 | beta | retaining | anti | 1uhv | β-xylosidase | Thermoanaerobacterium saccharolyticum | 2-F-xylosyl | Glu160 | Glu277 | [39] |
| GH42 | A | (β/α)8 | beta | retaining | anti | 1kwk | β-galactosidase | Thermus thermophylus A4 | d-galactose | Glu141 | Glu312 | [40] |
| GH44 | none | (β/α)8 | beta | retaining | anti | 2eqd | endoglucanase | Clostridium thermocellum | cellooctaose | Glu186 | Glu359 | [41] |
| GH45 | none | 6-strand. β-barrel | beta | inverting | syn | 4eng | endo-1,4-glucanase | Humicola insolens | cellohexaose | Asp121 | Asp10 | [42] |
| GH46 | I | α + β | beta | inverting | predicted syn by clan | see at GH24 | ||||||
| GH47 | none | (α/α)7 | alpha | inverting | anti | 1x9d | α-mannosidase I | Homo sapiens | Me-2-S-(α-Man)-2-thio-α-Man | Asp463 | Glu599 | [43], [44] |
| GH48 | M | (α/α)6 | beta | inverting | predicted anti by clan | see at GH8 | ||||||
| GH49 | N | β-helix | alpha | inverting | predicted anti by clan | see at GH28 | ||||||
| GH50 | A | (β/α)8 | beta | retaining | predicted anti by clan | see e.g. at GH1 | ||||||
| GH51 | A | (β/α)8 | alpha | retaining | anti | 1qw9 | α-l-arabino- furanosidase | Geobacillus stearothermophilus | PNP-l-arabino-furanoside | Glu175 | Glu294 | [45] |
| GH53 | A | (β/α)8 | beta | retaining | predicted anti by clan | see e.g. at GH1 | ||||||
| GH54 | none | β-sandwich | alpha | retaining | anti | 1wd4 | α-l-arabino- furanosidase B | Aspergillus kawachii | l-arabinofuranose | Asp297 | Glu221 | [46] |
| GH55 | none | β-helix | beta | inverting | anti | 3eqo | β-1,3-glucanase | Phanerochaete chrysosporium K-3 | d-gluconolacton | Glu633 | unknown | [47] |
| GH56 | none | (β/α)7 | beta | retaining | anti | 1fcv | hyaluronidase | Apis mellifera | (hyaluron.)4 | Glu113 | internal | [48] |
| GH57 | none | (β/α)7 | alpha | retaining | anti | 1kly | glucanotransferase | Thermococcus litoralis | acarbose | Asp214 | Glu123 | [49] |
| GH59 | A | (β/α)8 | beta | retaining | predicted anti by clan | see e.g. at GH1 | ||||||
| GH63 | G | (α/α)6 | alpha | inverting | predicted anti by clan | see at GH37 | ||||||
| GH65 | L | (α/α)6 | alpha | inverting | predicted syn by clan | see at GH15 | ||||||
| GH67 | none | (β/α)8 | alpha | inverting | syn | 1gql | α-glucuronidase | Cellvibrio japonicus Ueda107 | d-glucuronic acid | Glu292 | unknown | [50] |
| GH68 | J | 5-fold β-propeller | beta | retaining | anti | 1pt2 | levansucrase | Bacillus subtilis | sucrose | Glu342 | Asp86 | [51] |
| GH70 | H | (β/α)8 | alpha | retaining | predicted anti by clan | see e.g. at GH13 | ||||||
| GH72 | A | (β/α)8 | beta | retaining | anti | 2w62 | β-1,3-glucano- transferase | Saccharomyces cerevisiae S288C | laminaripentaose | Glu176 | Glu275 | [52] |
| GH74 | none | 7-fold β-propeller | beta | inverting | syn | 2ebs | cellobiohydrolase (OXG-RCBH) | Geotrichum sp. m128 | xyloglucan heptasaccharide | Asp465 | Asp35 | [53] |
| GH77 | H | (β/α)8 | alpha | retaining | anti | 1esw | amylomaltase | Thermus aquaticus | acarbose | Asp395 | Asp293 | [54] |
| GH79 | A | (β/α)8 | beta | retaining | predicted anti by clan | see e.g. at GH1 | ||||||
| GH80 | I | α + β | beta | inverting | predicted syn by clan | see at GH24 | ||||||
| GH83 | E | 6-fold β-propeller | alpha | retaining | predicted anti by clan | see e.g. at GH33 | ||||||
| GH84 | none | (β/α)8 | beta | retaining | anti | 2chn | β-N-acetyl- glucosaminidase | Bacteroides thetaiota- omicron VPI-5482 | NAG-thiazoline | Glu242 | internal | [55] |
| GH85 | K | (β/α)8 | beta | retaining | anti | 2w92 | endo-β-N-acetyl- glucosaminidase D | Streptococcus pneumoniae TIGR4 | NAG-thiazoline | Glu337 | internal | [56] |
| GH86 | A | (β/α)8 | beta | retaining | predicted anti by clan | see e.g. at GH1 | ||||||
| GH89 | none | (β/α)8 | alpha | retaining | anti | 2vcb | α-N-acetyl- glucosaminidase | Clostridium perfringens | PUGNAc | Glu483 | Glu601 | [57] |
| GH92 | none | (α/α)6 + β-sandw. | alpha | inverting | anti | 2ww1 | α-1,2-mannosidase | Bacteroides thetaiota- omicron VPI-5482 | thiomannobioside | Glu533 | Asp644 Asp642 | [58] |
| GH93 | E | 6-fold β-propeller | alpha | retaining | predicted anti by clan | see e.g. at GH33 | ||||||
| GH94 | none | (α/α)6 | beta | inverting | syn | 1v7x | chitobiose phosphorylase | Vibrio proteolyticus | GlcNAc | Asp492 | phosphate | [59] |
| GH95 | none | (α/α)6 | alpha | inverting | anti | 2ead | α-1,2-l-fucosidase | Bifidobacterium bifidum | Fuc-α-1,2-Gal | Glu566 | Asn423 Asp766 | [60] |
| GH97 | none | (β/α)8 | alpha | retaining + inverting | anti | 2zq0 | α-glucosidase | Bacteroides thetaiota- omicron VPI-5482 | acarbose | Glu532 | Glu508 | [61] |
| GH99 | none | (β/α)8 | alpha | retaining | anti | 4ad4 | endo-α-mannosidase | Bacteroides xylanisolvens | glucose-1,3-isofagomine and α-1,2- mannobiose | Glu336 | debated | [62] |
| GH102 | none | double-ψ β-barrel | beta | retaining | syn | 2pi8 | lytic transglycosylase A | Escherechia coli | chitohexaose | Asp308 | none | [63] |
| GH113 | A | (β/α)8 | beta | retaining | predicted anti by clan | see e.g. at GH1 |
References
Error fetching PMID 23137336:
Error fetching PMID 19733839:
Error fetching PMID 17002288:
Error fetching PMID 18976664:
Error fetching PMID 11709165:
Error fetching PMID 12595701:
Error fetching PMID 10508787:
Error fetching PMID 10200171:
Error fetching PMID 17666401:
Error fetching PMID 11884144:
Error fetching PMID 14756552:
Error fetching PMID 19279191:
Error fetching PMID 10220321:
Error fetching PMID 15364577:
Error fetching PMID 10331869:
Error fetching PMID 12741813:
Error fetching PMID 8679589:
Error fetching PMID 15062085:
Error fetching PMID 11560481:
Error fetching PMID 10884356:
Error fetching PMID 11518970:
Error fetching PMID 15299731:
Error fetching PMID 8259514:
Error fetching PMID 12203498:
Error fetching PMID 12005440:
Error fetching PMID 17397864:
Error fetching PMID 14715651:
Error fetching PMID 18036614:
Error fetching PMID 17335500:
Error fetching PMID 15130470:
Error fetching PMID 1438172:
Error fetching PMID 15491613:
Error fetching PMID 17455176:
Error fetching PMID 12960159:
Error fetching PMID 14659747:
Error fetching PMID 12215416:
Error fetching PMID 17905739:
Error fetching PMID 15299721:
Error fetching PMID 15713668:
Error fetching PMID 18619586:
Error fetching PMID 14517232:
Error fetching PMID 15292273:
Error fetching PMID 19193645:
Error fetching PMID 12618437:
Error fetching PMID 11937059:
Error fetching PMID 14517548:
Error fetching PMID 19097997:
Error fetching PMID 17498741:
Error fetching PMID 11082203:
Error fetching PMID 16565725:
Error fetching PMID 19181667:
Error fetching PMID 18443291:
Error fetching PMID 15274915:
Error fetching PMID 17459873:
Error fetching PMID 18981178:
Error fetching PMID 17502382:
Error fetching PMID 22219371:
-
Heightman, T.D. and Vasella, A.T. (1999) Recent Insights into Inhibition, Structure, and Mechanism of Configuration-Retaining Glycosidases. Angewandte Chemie-International Edition 38(6), 750-770. Article online.
- Error fetching PMID 15642336:
- Error fetching PMID 23137336:
-
Pérez S and Marchessault RH (1978) The exo-anomeric effect: experimental evidence from crystal structures. Carbohydr res 65, 114-120.
-
Cramer CJ, Truhlar DG and French AD (1997) Exo-anomeric effects on energies and geometries of different conformations of glucose and related systems in the gas phase and aqueous solution. Carbohydr res 298, 1-14.
- Error fetching PMID 19733839:
- Error fetching PMID 17002288:
- Error fetching PMID 18976664:
- Error fetching PMID 11709165:
- Error fetching PMID 12595701:
- Error fetching PMID 10508787:
- Error fetching PMID 10200171:
- Error fetching PMID 11884144:
- Error fetching PMID 14756552:
- Error fetching PMID 19279191:
- Error fetching PMID 10220321:
- Error fetching PMID 15364577:
- Error fetching PMID 10331869:
- Error fetching PMID 12741813:
- Error fetching PMID 8679589:
- Error fetching PMID 15062085:
- Error fetching PMID 11560481:
- Error fetching PMID 10884356:
- Error fetching PMID 11518970:
- Error fetching PMID 15299731:
- Error fetching PMID 8259514:
- Error fetching PMID 12203498:
- Error fetching PMID 12005440:
- Error fetching PMID 17397864:
- Error fetching PMID 14715651:
- Error fetching PMID 17666401:
- Error fetching PMID 18036614:
- Error fetching PMID 17335500:
- Error fetching PMID 15130470:
- Error fetching PMID 1438172:
- Error fetching PMID 15491613:
- Error fetching PMID 17455176:
- Error fetching PMID 12960159:
- Error fetching PMID 14659747:
- Error fetching PMID 12215416:
- Error fetching PMID 17905739:
- Error fetching PMID 15299721:
- Error fetching PMID 15713668:
- Error fetching PMID 18619586:
- Error fetching PMID 14517232:
- Error fetching PMID 15292273:
- Error fetching PMID 19193645:
-
Marković-Housley Z, Miglierini G, Soldatova L, Rizkallah PJ, Müller U, Schirmer T. Crystal structure of hyaluronidase, a major allergen of bee venom. Structure. 2000 Oct 15;8(10):1025-35.
Note: Due to a problem with PubMed data, this reference is not automatically formatted. Please see these links out: DOI:10.1016/S0969-2126(00)00511-6 PMID:11080624
- Error fetching PMID 12618437:
- Error fetching PMID 11937059:
- Error fetching PMID 14517548:
- Error fetching PMID 19097997:
- Error fetching PMID 17498741:
- Error fetching PMID 11082203:
- Error fetching PMID 16565725:
- Error fetching PMID 19181667:
- Error fetching PMID 18443291:
-
Zhu et al. (2010) Nature Chemical Biology in the press; DOI: 10.1038/nchembio.278 direct link.
- Error fetching PMID 15274915:
- Error fetching PMID 17459873:
- Error fetching PMID 18981178:
- Error fetching PMID 22219371:
- Error fetching PMID 17502382: