CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Glycoside Hydrolase Family 3"
Harry Brumer (talk | contribs) |
Harry Brumer (talk | contribs) |
||
| Line 29: | Line 29: | ||
== Substrate specificities == | == Substrate specificities == | ||
| − | |||
There are a very large number of enzymes in this family and most originate from microorganisms. Their classification is based largely on nucleotide and amino acid sequence similarities of the corresponding genes. Relatively few members of the enzyme family have been purified and characterised in detail. | There are a very large number of enzymes in this family and most originate from microorganisms. Their classification is based largely on nucleotide and amino acid sequence similarities of the corresponding genes. Relatively few members of the enzyme family have been purified and characterised in detail. | ||
| − | The family 3 enzymes have been classified as β-D-glucosidases, α-L-arabinofuranosidases, β-D-xylopyranosidases and N-acetyl-β-D-glucosaminidases | + | The family 3 enzymes have been classified as β-D-glucosidases, α-L-arabinofuranosidases, β-D-xylopyranosidases and N-acetyl-β-D-glucosaminidases <cite>Harvey2000</cite>. In many cases the enzymes have dual or broad substrate specificities with respect to monosaccharide residues, linkage position and chain length of the substrate. For example, there are several well characterized ‘bifunctional’ enzymes in the family that have both α-L-arabinofuranosidase and β-D-xylopyranosidase activity <cite>Lee2003</cite>. In another example, the family 3 β-D-glucosidases from barley, which are more precisely referred to as β-D-glucan glucohydrolases, are broad specificity exo-hydrolases that remove single glucosyl residues from the non-reducing ends of a range of β-D-glucans, β-D-oligoglucosides and aryl b-D-glucosides, including (1,3)-β-D-glucans, (1,4)-β-D-glucans, (1,3;1,4)-β-D-glucans and (1,6)-β-D-glucans, 4nitrophenyl-β-D-glucoside, certain cyanogenic β-D-glucosides and some β-D-oligoxyloglucosides <cite>Hrmova1998</cite>. |
| − | In contrast, family 3 N-acetyl-β-D-glucosaminidases (NagZ) are ‘monofunctional’ glycoside hydrolases that remove N-acetyl-β-D-glucosamine (GlcNAc) from glycoconjugates | + | In contrast, family 3 N-acetyl-β-D-glucosaminidases (NagZ) are ‘monofunctional’ glycoside hydrolases that remove N-acetyl-β-D-glucosamine (GlcNAc) from glycoconjugates <cite>Chitlaru1996</cite>. Highly conserved in Gram-negative bacteria, NagZ enzymes play an important role in peptidoglycan recycling by removing GlcNAc from 1,6-anhydroMurNAc-peptides <cite>Cheng2000</cite>, and this activity has been shown to mediate the induction of chromosomal AmpC beta-lactamase <cite>Votsch2000 Asgarali2009</cite>. |
== Kinetics and Mechanism == | == Kinetics and Mechanism == | ||
| − | + | Family 3 enzymes remove single glycosyl residues from the non-reducing ends of their substrates. Catalysis occurs via a double displacement mechanism and the β-anomeric configuration of the released glucose molecule is retained. The stereochemistry of the reaction has been determined experimentally for some family 3 enzymes. Detailed kinetic analyses are available for two purified barley β-D-glucan glucohydrolases and two barley ‘bifunctional’ α-L-arabinofuranosidase/β-D-xylopyranosidases <cite>Lee2003 Hrmova1998</cite>. Detailed kinetic data are also available for a N-acetyl-β-D-glucosaminidase from ''Vibrio furnisii'' (ExoII) <cite>Vocadlo2000</cite> | |
| − | Family 3 enzymes remove single glycosyl residues from the non-reducing ends of their substrates. Catalysis occurs via a double displacement mechanism and the β-anomeric configuration of the released glucose molecule is retained. The stereochemistry of the reaction has been determined experimentally for some family 3 enzymes. Detailed kinetic analyses are available for two purified barley β-D-glucan glucohydrolases and two barley ‘bifunctional’ α-L-arabinofuranosidase/β-D-xylopyranosidases | ||
== Catalytic Residues == | == Catalytic Residues == | ||
| + | The catalytic amino acid residues for the barley β-D-glucan glucohydrolases have been identified by chemical and three-dimensional structural procedures <cite>Vargese1999</cite>. The substrate-binding site consists of two glucosyl-binding subsites and the catalytic amino acid residues are located between these two subsites. In the plant family 3 β-D-glycosidases the catalytic nucleophile is Asp285, which is located in a highly conserved GFVISDW motif. The catalytic acid, E491, is highly conserved in plant family 3 enzymes but is more difficult to locate in more distantly related members of the family <cite>Harvey2000</cite>. The reaction sequence and mechanism have been defined for this enzyme using a range of synthetic inhibitors <cite>Hrmova2001</cite>. | ||
| − | + | The [[catalytic nucleophile]] for ''Vibrio furnisii'' ExoII (a NagZ) has been identified chemically as Asp242, which is conserved thought the family 3 NagZ enzymes <cite>Vocadlo2000</cite>. A [[general acid]] residue has not been identified for family 3 NagZ enzymes. | |
| − | |||
| − | The [[catalytic nucleophile]] for ''Vibrio furnisii'' ExoII (a NagZ) has been identified chemically as Asp242, which is conserved thought the family 3 NagZ enzymes | ||
== Three-dimensional structures == | == Three-dimensional structures == | ||
| − | The family 3 β-D-glycosidases are globular monomeric enzymes of molecular mass around 60-70 kDa. The 3D structure of the β-D-glucan glucohydrolase isoenzyme ExoII from barley, determined by X-ray crystallography to 2.2 Å resolution, shows a two-domain, globular protein of 605 amino acid residues that is N-glycosylated at three sites | + | The family 3 β-D-glycosidases are globular monomeric enzymes of molecular mass around 60-70 kDa. The 3D structure of the β-D-glucan glucohydrolase isoenzyme ExoII from barley, determined by X-ray crystallography to 2.2 Å resolution, shows a two-domain, globular protein of 605 amino acid residues that is N-glycosylated at three sites <cite>Vargese1999</cite>. The two domains are connected by a 16-amino acid helix-like linker. The first 357 residues constitute a (β/α)<sub>8</sub> TIM barrel domain. The second domain consists of residues 374 to 559 arranged in a six-stranded β-sandwich, which contains a β-sheet of five parallel β-strands and one antiparallel β-strand, with 3 α-helices on either side of the sheet. A long antiparallel loop of 42 amino acid residues is found at the COOH-terminus of the enzyme. In some bacterial GH3 enzymes the order of the domains is reversed <cite>Harvey2000</cite>. |
| − | The active site of the barley β-D-glucan glucohydrolase consists of a relatively shallow substrate-binding pocket that is located at the interface of the two domains of the enzyme | + | The active site of the barley β-D-glucan glucohydrolase consists of a relatively shallow substrate-binding pocket that is located at the interface of the two domains of the enzyme <cite>Vargese1999</cite>. The active site pocket can accommodate the two glucosyl residues at the non-reducing terminus of the substrate and aligns the non-reducing terminal glycosidic linkage of the substrate with the catalytic amino acid residues Asp285 and Glu491. Thus, the catalytic amino acid residues are located on domains 1 and 2, respectively. |
| − | The broad specificity of the barley β-D-glucan glucohydrolase can be rationalized from the X-ray crystallographic data and from molecular modelling of enzyme-substrate complexes | + | The broad specificity of the barley β-D-glucan glucohydrolase can be rationalized from the X-ray crystallographic data and from molecular modelling of enzyme-substrate complexes <cite>Vargese1999 Hrmova2002</cite>. The glucosyl residue occupying binding subsite –1 is tightly locked into a relatively fixed position through interactions with six amino acid residues near the bottom of the shallow active site pocket. In contrast, the glucosyl residue at subsite +1 is located between two tryptophan residues at the entrance of the pocket, where it is less tightly constrained. The flexibility of binding at subsite +1, coupled with the projection of the remainder of bound substrate away from the enzyme’s surface, means that the overall active site is largely independent of substrate conformation and will therefore accommodate a range of substrates in which the spatial dispositions of adjacent β-D-glucosyl residues vary as a result of glycosidic linkages between different C atoms of the adjacent β-D-glucosyl residues <cite>Hrmova2002</cite>. |
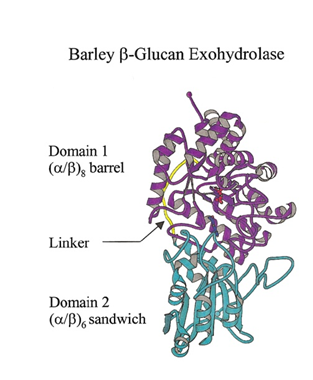
[[File:GH3_Fig_1.png]] | [[File:GH3_Fig_1.png]] | ||
| − | Figure 1. Ribbon representation of barley β-glucan exohydrolase isoenzyme ExoI. Domain 1, domain 2, and the linker region of the enzyme are coloured in magenta, cyan, and yellow, respectively. Figure from | + | Figure 1. Ribbon representation of barley β-glucan exohydrolase isoenzyme ExoI. Domain 1, domain 2, and the linker region of the enzyme are coloured in magenta, cyan, and yellow, respectively. Figure from <cite>Harvey2000</cite>. |
| − | Family 3 NagZ enzymes are also globular, yet have a mass of ~ 36 kDa, which is a distinctive feature of NagZ enzymes from others within the family. NagZ from ''Vibrio cholerae'' has been determined in complex with GlcNAc (PDB ID: 1Y65) and with the N-acetyl-β-glucosaminidase inhibitor PUGNAc | + | Family 3 NagZ enzymes are also globular, yet have a mass of ~ 36 kDa, which is a distinctive feature of NagZ enzymes from others within the family. NagZ from ''Vibrio cholerae'' has been determined in complex with GlcNAc (PDB ID: 1Y65) and with the N-acetyl-β-glucosaminidase inhibitor PUGNAc <cite>Stubbs2007</cite> and NagZ selective PUGNAc derivatives <cite>Balcewich2009</cite>. The enzyme is comprised of 340 amino acids and adopts a (β/α)<sub>8</sub> TIM barrel fold. The active site pocket is shallow and accommodates the 2-N-acetyl group of a terminal GlcNAc sugar in a solvent accessible groove alongside the binding site for the pyranose ring. This active site architecture is different from the active site architectures the functionally related family 20 N-acetyl-β-hexosaminidases and family 84 O-GlcNAcases. These latter families, which also remove β-1,4-linked GlcNAc residues from glycoconjugates, use a mechanism involving [[neighboring group participation]] wherein the carbonyl oxygen of the 2-acetamido group of the terminal GlcNAc acts as a nucleophile, yielding an [[oxazolinium ion]] [[intermediate]] <cite>Tews1996 Mark2001 Dennis2006</cite>. Thus, unlike family 3 NagZ enzymes, family 20 and 84 enzymes do not possess an enzymic [[catalytic nucleophile]]; however, they do have an appropriately positioned catalytic acid residue. Together, these mechanistic differences have allowed for the development of 2-N-acyl derivatives of PUGNAc that are selective for family 3 NagZ over family 20 and 84 enzymes <cite>Stubbs2007 Balcewich2009</cite>. |
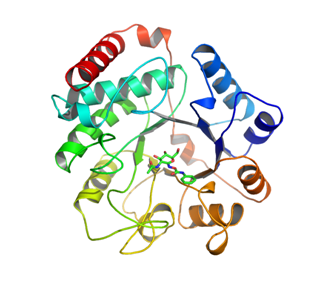
[[File:GH3_Fig_2.png]] | [[File:GH3_Fig_2.png]] | ||
| − | Figure 2 : NagZ from ''Vibrio cholerae'' in complex with PUGNAc (PDB ID: 2OXN) | + | Figure 2 : NagZ from ''Vibrio cholerae'' in complex with PUGNAc (PDB ID: 2OXN) <cite>Stubbs2007</cite>. NagZ enzymes are single domain proteins that adopt a TIM barrel fold. Active site residues are located within the loops that extend from the C-termini of the strands of the β-barrel. |
== Family Firsts == | == Family Firsts == | ||
| − | + | ;First 3D Structure: Barley <cite>Vargese1999</cite>. | |
| − | First 3D Structure | + | ;First Catalytic Residues: Barley <cite>Vargese1999</cite>. |
| − | |||
| − | Barley | ||
| − | |||
| − | |||
| − | First Catalytic Residues | ||
| − | |||
| − | Barley | ||
== References == | == References == | ||
| − | + | <biblio> | |
| − | + | #Harvey2000 pmid=10966578 | |
| − | + | #Lee2003 pmid=12464603 | |
2. Lee, R.C., Hrmova, M., Burton, R.A., Lahnstein, J. and Fincher, G.B. (2003) Bifunctional Family 3 Glycoside Hydrolases from Barley with α-L-Arabinofuranosidase and β-D-Xylosidase Activity: Characterization, Primary Structures and COOH-terminal processing. J. Biol. Chem. 278, 5377-5387. | 2. Lee, R.C., Hrmova, M., Burton, R.A., Lahnstein, J. and Fincher, G.B. (2003) Bifunctional Family 3 Glycoside Hydrolases from Barley with α-L-Arabinofuranosidase and β-D-Xylosidase Activity: Characterization, Primary Structures and COOH-terminal processing. J. Biol. Chem. 278, 5377-5387. | ||
| − | + | #Hrmova1998 Hrmova, M. and Fincher, G.B. (1998) Barley ß-D-glucan exohydrolases. Substrate specificity and kinetic properties. Carbohydr. Res. 305, 209-221. | |
| − | + | #Chitlaru1996 pmid=8969206 | |
| − | |||
4. Chitlaru, E. & Roseman, S. (1996). Molecular cloning and characterization of a novel beta-N-acetyl-D- glucosaminidase from Vibrio furnissii. J Biol Chem 271, 33433-9. | 4. Chitlaru, E. & Roseman, S. (1996). Molecular cloning and characterization of a novel beta-N-acetyl-D- glucosaminidase from Vibrio furnissii. J Biol Chem 271, 33433-9. | ||
| − | + | #Cheng2000 pmid=10940025 | |
5. Cheng, Q., Li, H., Merdek, K. & Park, J. T. (2000). Molecular characterization of the beta-N-acetylglucosaminidase of Escherichia coli and its role in cell wall recycling. J Bacteriol 182, 4836-40. | 5. Cheng, Q., Li, H., Merdek, K. & Park, J. T. (2000). Molecular characterization of the beta-N-acetylglucosaminidase of Escherichia coli and its role in cell wall recycling. J Bacteriol 182, 4836-40. | ||
| − | + | #Votsch2000 pmid=10978324 | |
6. Votsch, W. & Templin, M. F. (2000). Characterization of a beta-N-acetylglucosaminidase of Escherichia coli and elucidation of its role in muropeptide recycling and beta-lactamase induction. J Biol Chem 275, 39032-8 | 6. Votsch, W. & Templin, M. F. (2000). Characterization of a beta-N-acetylglucosaminidase of Escherichia coli and elucidation of its role in muropeptide recycling and beta-lactamase induction. J Biol Chem 275, 39032-8 | ||
| − | + | #Asgarali2009 pmid=19273679 | |
7. Asgarali, A., Stubbs, K. A., Oliver, A., Vocadlo, D. J. & Mark, B. L. (2009). Inactivation of the glycoside hydrolase NagZ attenuates antipseudomonal beta-lactam resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 53, 2274-82. | 7. Asgarali, A., Stubbs, K. A., Oliver, A., Vocadlo, D. J. & Mark, B. L. (2009). Inactivation of the glycoside hydrolase NagZ attenuates antipseudomonal beta-lactam resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 53, 2274-82. | ||
| − | + | #Vocadlo2000 pmid=10625486 | |
8. Vocadlo, D. J., Mayer, C., He, S. & Withers, S. G. (2000). Mechanism of action and identification of Asp242 as the catalytic nucleophile of Vibrio furnisii N-acetyl-beta-D-glucosaminidase using 2-acetamido-2-deoxy-5-fluoro-alpha-L-idopyranosyl fluoride. Biochemistry 39, 117-26. | 8. Vocadlo, D. J., Mayer, C., He, S. & Withers, S. G. (2000). Mechanism of action and identification of Asp242 as the catalytic nucleophile of Vibrio furnisii N-acetyl-beta-D-glucosaminidase using 2-acetamido-2-deoxy-5-fluoro-alpha-L-idopyranosyl fluoride. Biochemistry 39, 117-26. | ||
| − | + | #Vargese1999 pmid=10368285 | |
9. Varghese, J.N., Hrmova, M. and Fincher, G.B. (1999) Three-dimensional structure of a barley b-D-glucan exohydrolase; a family 3 hydrolase. Structure 7,179-190. | 9. Varghese, J.N., Hrmova, M. and Fincher, G.B. (1999) Three-dimensional structure of a barley b-D-glucan exohydrolase; a family 3 hydrolase. Structure 7,179-190. | ||
| − | + | #Hrmova2001 pmid=11709165 | |
10. Hrmova, M., Varghese, J.N., De Gori, R., Smith, B.J., Driguez, H. and Fincher, G.B. (2001) Catalytic Mechanisms and Reaction Intermediates along the Hydrolytic Pathway of a Plant β-d-Glucan Glucohydrolase. Structure 9, 1005-1016. | 10. Hrmova, M., Varghese, J.N., De Gori, R., Smith, B.J., Driguez, H. and Fincher, G.B. (2001) Catalytic Mechanisms and Reaction Intermediates along the Hydrolytic Pathway of a Plant β-d-Glucan Glucohydrolase. Structure 9, 1005-1016. | ||
| − | + | #Hrmova2002 pmid=12034895 | |
11. Hrmova M, De Gori R, Smith BJ, Fairweather JK, Driguez H, Varghese JN, Fincher GB (2002) Structural basis for broad substrate specificity in higher plant β-d-glucan glucohydrolases. The Plant Cell 14, 1033-1052. | 11. Hrmova M, De Gori R, Smith BJ, Fairweather JK, Driguez H, Varghese JN, Fincher GB (2002) Structural basis for broad substrate specificity in higher plant β-d-glucan glucohydrolases. The Plant Cell 14, 1033-1052. | ||
| − | + | #Stubbs2007 pmid=17439950 | |
12. Stubbs, K. A., Balcewich, M., Mark, B. L. & Vocadlo, D. J. (2007). Small molecule inhibitors of a glycoside hydrolase attenuate inducible AmpC-mediated beta-lactam resistance. J Biol Chem 282, 21382-91. | 12. Stubbs, K. A., Balcewich, M., Mark, B. L. & Vocadlo, D. J. (2007). Small molecule inhibitors of a glycoside hydrolase attenuate inducible AmpC-mediated beta-lactam resistance. J Biol Chem 282, 21382-91. | ||
| − | + | #Balcewich2009 pmid=19499593 | |
13. Balcewich, M. D., Stubbs, K. A., He, Y., James, T. W., Davies, G. J., Vocadlo, D. J. & Mark, B. L. (2009). Insight into a strategy for attenuating AmpC-mediated beta-lactam resistance: structural basis for selective inhibition of the glycoside hydrolase NagZ. Protein Sci 18, 1541-51. | 13. Balcewich, M. D., Stubbs, K. A., He, Y., James, T. W., Davies, G. J., Vocadlo, D. J. & Mark, B. L. (2009). Insight into a strategy for attenuating AmpC-mediated beta-lactam resistance: structural basis for selective inhibition of the glycoside hydrolase NagZ. Protein Sci 18, 1541-51. | ||
| − | + | #Tews1996 pmid=8673609 | |
14. Tews, I., Perrakis, A., Oppenheim, A., Dauter, Z., Wilson, K. S. & Vorgias, C. E. (1996). Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nat Struct Biol 3, 638-48. | 14. Tews, I., Perrakis, A., Oppenheim, A., Dauter, Z., Wilson, K. S. & Vorgias, C. E. (1996). Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nat Struct Biol 3, 638-48. | ||
| − | + | #Mark2001 pmid=11124970 | |
15. Mark, B. L., Vocadlo, D. J., Knapp, S., Triggs-Raine, B. L., Withers, S. G. & James, M. N. (2001). Crystallographic evidence for substrate-assisted catalysis in a bacterial beta-hexosaminidase. J Biol Chem 276, 10330-7. | 15. Mark, B. L., Vocadlo, D. J., Knapp, S., Triggs-Raine, B. L., Withers, S. G. & James, M. N. (2001). Crystallographic evidence for substrate-assisted catalysis in a bacterial beta-hexosaminidase. J Biol Chem 276, 10330-7. | ||
| − | + | #Dennis2006 pmid=16565725 | |
16. Dennis, R. J., Taylor, E. J., Macauley, M. S., Stubbs, K. A., Turkenburg, J. P., Hart, S. J., Black, G. N., Vocadlo, D. J. & Davies, G. J. (2006). Structure and mechanism of a bacterial beta-glucosaminidase having O-GlcNAcase activity. Nat Struct Mol Biol 13, 365-71. | 16. Dennis, R. J., Taylor, E. J., Macauley, M. S., Stubbs, K. A., Turkenburg, J. P., Hart, S. J., Black, G. N., Vocadlo, D. J. & Davies, G. J. (2006). Structure and mechanism of a bacterial beta-glucosaminidase having O-GlcNAcase activity. Nat Struct Mol Biol 13, 365-71. | ||
[[Category:Glycoside Hydrolase Families|GH003]] | [[Category:Glycoside Hydrolase Families|GH003]] | ||
Revision as of 02:02, 22 June 2010
This page has been approved by the Responsible Curator as essentially complete. CAZypedia is a living document, so further improvement of this page is still possible. If you would like to suggest an addition or correction, please contact the page's Responsible Curator directly by e-mail.
- Authors: ^^^Geoff Fincher^^^ and ^^^Brian Mark^^^
- Responsible Curator: ^^^Bernard Henrissat^^^
| Glycoside Hydrolase Family GH3 | |
| Clan | none |
| Mechanism | retaining |
| Active site residues | known |
| CAZy DB link | |
| http://www.cazy.org/GH3.html | |
Substrate specificities
There are a very large number of enzymes in this family and most originate from microorganisms. Their classification is based largely on nucleotide and amino acid sequence similarities of the corresponding genes. Relatively few members of the enzyme family have been purified and characterised in detail.
The family 3 enzymes have been classified as β-D-glucosidases, α-L-arabinofuranosidases, β-D-xylopyranosidases and N-acetyl-β-D-glucosaminidases [1]. In many cases the enzymes have dual or broad substrate specificities with respect to monosaccharide residues, linkage position and chain length of the substrate. For example, there are several well characterized ‘bifunctional’ enzymes in the family that have both α-L-arabinofuranosidase and β-D-xylopyranosidase activity [2]. In another example, the family 3 β-D-glucosidases from barley, which are more precisely referred to as β-D-glucan glucohydrolases, are broad specificity exo-hydrolases that remove single glucosyl residues from the non-reducing ends of a range of β-D-glucans, β-D-oligoglucosides and aryl b-D-glucosides, including (1,3)-β-D-glucans, (1,4)-β-D-glucans, (1,3;1,4)-β-D-glucans and (1,6)-β-D-glucans, 4nitrophenyl-β-D-glucoside, certain cyanogenic β-D-glucosides and some β-D-oligoxyloglucosides [3].
In contrast, family 3 N-acetyl-β-D-glucosaminidases (NagZ) are ‘monofunctional’ glycoside hydrolases that remove N-acetyl-β-D-glucosamine (GlcNAc) from glycoconjugates [4]. Highly conserved in Gram-negative bacteria, NagZ enzymes play an important role in peptidoglycan recycling by removing GlcNAc from 1,6-anhydroMurNAc-peptides [5], and this activity has been shown to mediate the induction of chromosomal AmpC beta-lactamase [6, 7].
Kinetics and Mechanism
Family 3 enzymes remove single glycosyl residues from the non-reducing ends of their substrates. Catalysis occurs via a double displacement mechanism and the β-anomeric configuration of the released glucose molecule is retained. The stereochemistry of the reaction has been determined experimentally for some family 3 enzymes. Detailed kinetic analyses are available for two purified barley β-D-glucan glucohydrolases and two barley ‘bifunctional’ α-L-arabinofuranosidase/β-D-xylopyranosidases [2, 3]. Detailed kinetic data are also available for a N-acetyl-β-D-glucosaminidase from Vibrio furnisii (ExoII) [8]
Catalytic Residues
The catalytic amino acid residues for the barley β-D-glucan glucohydrolases have been identified by chemical and three-dimensional structural procedures [9]. The substrate-binding site consists of two glucosyl-binding subsites and the catalytic amino acid residues are located between these two subsites. In the plant family 3 β-D-glycosidases the catalytic nucleophile is Asp285, which is located in a highly conserved GFVISDW motif. The catalytic acid, E491, is highly conserved in plant family 3 enzymes but is more difficult to locate in more distantly related members of the family [1]. The reaction sequence and mechanism have been defined for this enzyme using a range of synthetic inhibitors [10].
The catalytic nucleophile for Vibrio furnisii ExoII (a NagZ) has been identified chemically as Asp242, which is conserved thought the family 3 NagZ enzymes [8]. A general acid residue has not been identified for family 3 NagZ enzymes.
Three-dimensional structures
The family 3 β-D-glycosidases are globular monomeric enzymes of molecular mass around 60-70 kDa. The 3D structure of the β-D-glucan glucohydrolase isoenzyme ExoII from barley, determined by X-ray crystallography to 2.2 Å resolution, shows a two-domain, globular protein of 605 amino acid residues that is N-glycosylated at three sites [9]. The two domains are connected by a 16-amino acid helix-like linker. The first 357 residues constitute a (β/α)8 TIM barrel domain. The second domain consists of residues 374 to 559 arranged in a six-stranded β-sandwich, which contains a β-sheet of five parallel β-strands and one antiparallel β-strand, with 3 α-helices on either side of the sheet. A long antiparallel loop of 42 amino acid residues is found at the COOH-terminus of the enzyme. In some bacterial GH3 enzymes the order of the domains is reversed [1].
The active site of the barley β-D-glucan glucohydrolase consists of a relatively shallow substrate-binding pocket that is located at the interface of the two domains of the enzyme [9]. The active site pocket can accommodate the two glucosyl residues at the non-reducing terminus of the substrate and aligns the non-reducing terminal glycosidic linkage of the substrate with the catalytic amino acid residues Asp285 and Glu491. Thus, the catalytic amino acid residues are located on domains 1 and 2, respectively.
The broad specificity of the barley β-D-glucan glucohydrolase can be rationalized from the X-ray crystallographic data and from molecular modelling of enzyme-substrate complexes [9, 11]. The glucosyl residue occupying binding subsite –1 is tightly locked into a relatively fixed position through interactions with six amino acid residues near the bottom of the shallow active site pocket. In contrast, the glucosyl residue at subsite +1 is located between two tryptophan residues at the entrance of the pocket, where it is less tightly constrained. The flexibility of binding at subsite +1, coupled with the projection of the remainder of bound substrate away from the enzyme’s surface, means that the overall active site is largely independent of substrate conformation and will therefore accommodate a range of substrates in which the spatial dispositions of adjacent β-D-glucosyl residues vary as a result of glycosidic linkages between different C atoms of the adjacent β-D-glucosyl residues [11].
Figure 1. Ribbon representation of barley β-glucan exohydrolase isoenzyme ExoI. Domain 1, domain 2, and the linker region of the enzyme are coloured in magenta, cyan, and yellow, respectively. Figure from [1].
Family 3 NagZ enzymes are also globular, yet have a mass of ~ 36 kDa, which is a distinctive feature of NagZ enzymes from others within the family. NagZ from Vibrio cholerae has been determined in complex with GlcNAc (PDB ID: 1Y65) and with the N-acetyl-β-glucosaminidase inhibitor PUGNAc [12] and NagZ selective PUGNAc derivatives [13]. The enzyme is comprised of 340 amino acids and adopts a (β/α)8 TIM barrel fold. The active site pocket is shallow and accommodates the 2-N-acetyl group of a terminal GlcNAc sugar in a solvent accessible groove alongside the binding site for the pyranose ring. This active site architecture is different from the active site architectures the functionally related family 20 N-acetyl-β-hexosaminidases and family 84 O-GlcNAcases. These latter families, which also remove β-1,4-linked GlcNAc residues from glycoconjugates, use a mechanism involving neighboring group participation wherein the carbonyl oxygen of the 2-acetamido group of the terminal GlcNAc acts as a nucleophile, yielding an oxazolinium ion intermediate [14, 15, 16]. Thus, unlike family 3 NagZ enzymes, family 20 and 84 enzymes do not possess an enzymic catalytic nucleophile; however, they do have an appropriately positioned catalytic acid residue. Together, these mechanistic differences have allowed for the development of 2-N-acyl derivatives of PUGNAc that are selective for family 3 NagZ over family 20 and 84 enzymes [12, 13].
Figure 2 : NagZ from Vibrio cholerae in complex with PUGNAc (PDB ID: 2OXN) [12]. NagZ enzymes are single domain proteins that adopt a TIM barrel fold. Active site residues are located within the loops that extend from the C-termini of the strands of the β-barrel.
Family Firsts
References
<biblio>
- Harvey2000 pmid=10966578
- Lee2003 pmid=12464603
2. Lee, R.C., Hrmova, M., Burton, R.A., Lahnstein, J. and Fincher, G.B. (2003) Bifunctional Family 3 Glycoside Hydrolases from Barley with α-L-Arabinofuranosidase and β-D-Xylosidase Activity: Characterization, Primary Structures and COOH-terminal processing. J. Biol. Chem. 278, 5377-5387.
- Hrmova1998 Hrmova, M. and Fincher, G.B. (1998) Barley ß-D-glucan exohydrolases. Substrate specificity and kinetic properties. Carbohydr. Res. 305, 209-221.
- Chitlaru1996 pmid=8969206
4. Chitlaru, E. & Roseman, S. (1996). Molecular cloning and characterization of a novel beta-N-acetyl-D- glucosaminidase from Vibrio furnissii. J Biol Chem 271, 33433-9.
- Cheng2000 pmid=10940025
5. Cheng, Q., Li, H., Merdek, K. & Park, J. T. (2000). Molecular characterization of the beta-N-acetylglucosaminidase of Escherichia coli and its role in cell wall recycling. J Bacteriol 182, 4836-40.
- Votsch2000 pmid=10978324
6. Votsch, W. & Templin, M. F. (2000). Characterization of a beta-N-acetylglucosaminidase of Escherichia coli and elucidation of its role in muropeptide recycling and beta-lactamase induction. J Biol Chem 275, 39032-8
- Asgarali2009 pmid=19273679
7. Asgarali, A., Stubbs, K. A., Oliver, A., Vocadlo, D. J. & Mark, B. L. (2009). Inactivation of the glycoside hydrolase NagZ attenuates antipseudomonal beta-lactam resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 53, 2274-82.
- Vocadlo2000 pmid=10625486
8. Vocadlo, D. J., Mayer, C., He, S. & Withers, S. G. (2000). Mechanism of action and identification of Asp242 as the catalytic nucleophile of Vibrio furnisii N-acetyl-beta-D-glucosaminidase using 2-acetamido-2-deoxy-5-fluoro-alpha-L-idopyranosyl fluoride. Biochemistry 39, 117-26.
- Vargese1999 pmid=10368285
9. Varghese, J.N., Hrmova, M. and Fincher, G.B. (1999) Three-dimensional structure of a barley b-D-glucan exohydrolase; a family 3 hydrolase. Structure 7,179-190.
- Hrmova2001 pmid=11709165
10. Hrmova, M., Varghese, J.N., De Gori, R., Smith, B.J., Driguez, H. and Fincher, G.B. (2001) Catalytic Mechanisms and Reaction Intermediates along the Hydrolytic Pathway of a Plant β-d-Glucan Glucohydrolase. Structure 9, 1005-1016.
- Hrmova2002 pmid=12034895
11. Hrmova M, De Gori R, Smith BJ, Fairweather JK, Driguez H, Varghese JN, Fincher GB (2002) Structural basis for broad substrate specificity in higher plant β-d-glucan glucohydrolases. The Plant Cell 14, 1033-1052.
- Stubbs2007 pmid=17439950
12. Stubbs, K. A., Balcewich, M., Mark, B. L. & Vocadlo, D. J. (2007). Small molecule inhibitors of a glycoside hydrolase attenuate inducible AmpC-mediated beta-lactam resistance. J Biol Chem 282, 21382-91.
- Balcewich2009 pmid=19499593
13. Balcewich, M. D., Stubbs, K. A., He, Y., James, T. W., Davies, G. J., Vocadlo, D. J. & Mark, B. L. (2009). Insight into a strategy for attenuating AmpC-mediated beta-lactam resistance: structural basis for selective inhibition of the glycoside hydrolase NagZ. Protein Sci 18, 1541-51.
- Tews1996 pmid=8673609
14. Tews, I., Perrakis, A., Oppenheim, A., Dauter, Z., Wilson, K. S. & Vorgias, C. E. (1996). Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nat Struct Biol 3, 638-48.
- Mark2001 pmid=11124970
15. Mark, B. L., Vocadlo, D. J., Knapp, S., Triggs-Raine, B. L., Withers, S. G. & James, M. N. (2001). Crystallographic evidence for substrate-assisted catalysis in a bacterial beta-hexosaminidase. J Biol Chem 276, 10330-7.
- Dennis2006 pmid=16565725
16. Dennis, R. J., Taylor, E. J., Macauley, M. S., Stubbs, K. A., Turkenburg, J. P., Hart, S. J., Black, G. N., Vocadlo, D. J. & Davies, G. J. (2006). Structure and mechanism of a bacterial beta-glucosaminidase having O-GlcNAcase activity. Nat Struct Mol Biol 13, 365-71.