CAZypedia needs your help! We have many unassigned GH, PL, CE, AA, GT, and CBM pages in need of Authors and Responsible Curators.
Scientists at all career stages, including students, are welcome to contribute to CAZypedia. Read more here, and in the 10th anniversary article in Glycobiology.
New to the CAZy classification? Read this first.
*
Consider attending the 15th Carbohydrate Bioengineering Meeting in Ghent, 5-8 May 2024.
Difference between revisions of "Glycoside Hydrolase Family 73"
| Line 37: | Line 37: | ||
== Three-dimensional structures == | == Three-dimensional structures == | ||
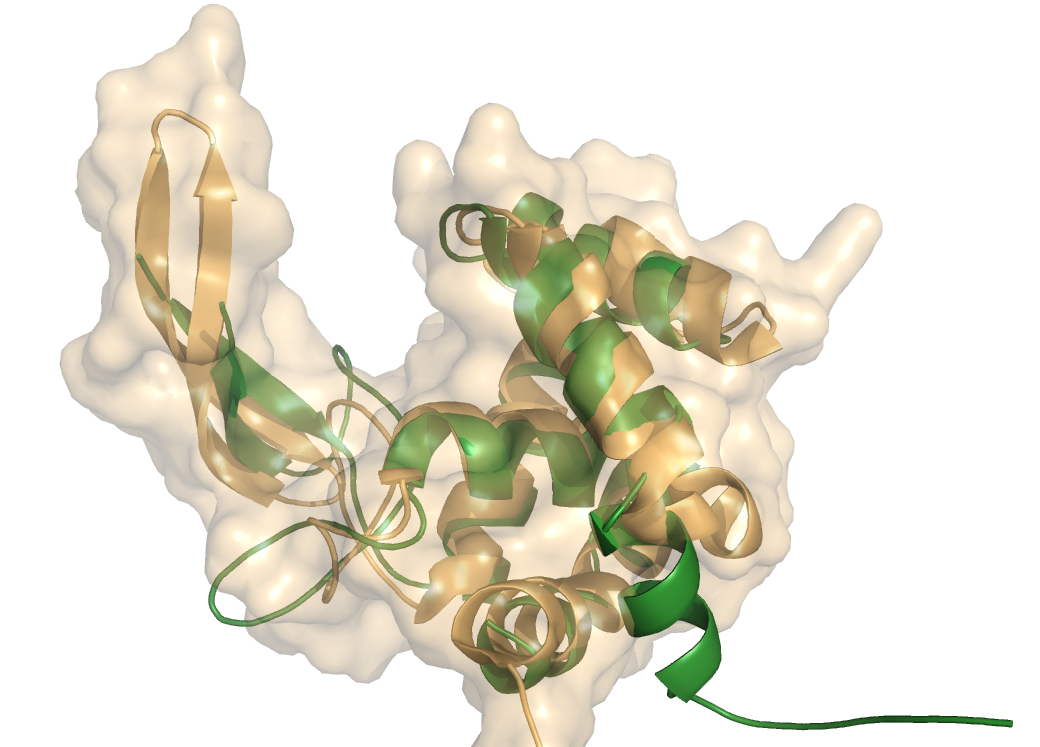
| − | [[Image:auto-flgjSURF.jpg|frame|right|'''Figure 1.''' Ribbon diagram of Auto structure (orange)and its surface superimposed on FlgJ structure (green). The (α+β) fold forms a deep substrate binding groove | + | [[Image:auto-flgjSURF.jpg|150px|frame|right|'''Figure 1.''' Ribbon diagram of Auto structure (orange) and its surface, superimposed on FlgJ structure (green). The (α+β) fold forms a deep substrate binding groove.]] |
crystal structure of GH73 are available and have been coincidently reported, FlgJ from ''Sphingomonas sp.'' (SPH1045-C) <cite>1</cite> and Auto a virulence associated peptigoglycan hydrolase from ''Listeria monocytogenes'' <cite>2</cite>. A structure for a catalytic mutant (E185A) of FlgJ has been solved by Maruyama et al <cite>3</cite> but doesn’t show any conformational changes. The two GH73 show the same fold, with two subdomains consisting of a β-lobe and an α-lobe that together create an extended substrate binding groove. With a typical lysozyme (α+β) fold, the catalytic domain of Auto is structurally related to the catalytic domain of Slt70 from ''E. coli'' <cite>4</cite>, the family [[GH19]] chitinases and goose egg-white lysozyme (GEWL, [[GH23]])<cite>5</cite>. FlgJ is structurally related to a peptidoglycan degrading enzyme from the bacteriophage phi 29 <cite>6</cite> and also to family [[GH22]] and [[GH23]] lysozymes. | crystal structure of GH73 are available and have been coincidently reported, FlgJ from ''Sphingomonas sp.'' (SPH1045-C) <cite>1</cite> and Auto a virulence associated peptigoglycan hydrolase from ''Listeria monocytogenes'' <cite>2</cite>. A structure for a catalytic mutant (E185A) of FlgJ has been solved by Maruyama et al <cite>3</cite> but doesn’t show any conformational changes. The two GH73 show the same fold, with two subdomains consisting of a β-lobe and an α-lobe that together create an extended substrate binding groove. With a typical lysozyme (α+β) fold, the catalytic domain of Auto is structurally related to the catalytic domain of Slt70 from ''E. coli'' <cite>4</cite>, the family [[GH19]] chitinases and goose egg-white lysozyme (GEWL, [[GH23]])<cite>5</cite>. FlgJ is structurally related to a peptidoglycan degrading enzyme from the bacteriophage phi 29 <cite>6</cite> and also to family [[GH22]] and [[GH23]] lysozymes. | ||
Revision as of 08:47, 23 July 2010
This page is currently under construction. This means that the Responsible Curator has deemed that the page's content is not quite up to CAZypedia's standards for full public consumption. All information should be considered to be under revision and may be subject to major changes.
- Author: ^^^Florence Vincent^^^
- Responsible Curator: ^^^Bernard Henrissat^^^
| Glycoside Hydrolase Family GH73 | |
| Clan | none, α+β "lysozyme fold" |
| Mechanism | not known |
| Active site residues | partially known |
| CAZy DB link | |
| http://www.cazy.org/GH73.html | |
Substrate specificities
Content is to be added here.
Kinetics and Mechanism
Content is to be added here.
Catalytic Residues
Content is to be added here.
Three-dimensional structures
crystal structure of GH73 are available and have been coincidently reported, FlgJ from Sphingomonas sp. (SPH1045-C) [1] and Auto a virulence associated peptigoglycan hydrolase from Listeria monocytogenes [2]. A structure for a catalytic mutant (E185A) of FlgJ has been solved by Maruyama et al [3] but doesn’t show any conformational changes. The two GH73 show the same fold, with two subdomains consisting of a β-lobe and an α-lobe that together create an extended substrate binding groove. With a typical lysozyme (α+β) fold, the catalytic domain of Auto is structurally related to the catalytic domain of Slt70 from E. coli [4], the family GH19 chitinases and goose egg-white lysozyme (GEWL, GH23)[5]. FlgJ is structurally related to a peptidoglycan degrading enzyme from the bacteriophage phi 29 [6] and also to family GH22 and GH23 lysozymes.
Family Firsts
- First stereochemistry determination
- Cite some reference here, with a short (1-2 sentence) explanation
- First catalytic nucleophile identification
- Cite some reference here, with a short (1-2 sentence) explanation
- First general acid/base residue identification
- Cite some reference here, with a short (1-2 sentence) explanation
- First 3-D structure
- Cite some reference here, with a short (1-2 sentence) explanation
References
- Hashimoto W, Ochiai A, Momma K, Itoh T, Mikami B, Maruyama Y, and Murata K. (2009). Crystal structure of the glycosidase family 73 peptidoglycan hydrolase FlgJ. Biochem Biophys Res Commun. 2009;381(1):16-21. DOI:10.1016/j.bbrc.2009.01.186 |
- Bublitz M, Polle L, Holland C, Heinz DW, Nimtz M, and Schubert WD. (2009). Structural basis for autoinhibition and activation of Auto, a virulence-associated peptidoglycan hydrolase of Listeria monocytogenes. Mol Microbiol. 2009;71(6):1509-22. DOI:10.1111/j.1365-2958.2009.06619.x |
- Maruyama Y, Ochiai A, Itoh T, Mikami B, Hashimoto W, and Murata K. (2010). Mutational studies of the peptidoglycan hydrolase FlgJ of Sphingomonas sp. strain A1. J Basic Microbiol. 2010;50(4):311-7. DOI:10.1002/jobm.200900249 |
- van Asselt EJ, Thunnissen AM, and Dijkstra BW. (1999). High resolution crystal structures of the Escherichia coli lytic transglycosylase Slt70 and its complex with a peptidoglycan fragment. J Mol Biol. 1999;291(4):877-98. DOI:10.1006/jmbi.1999.3013 |
- Weaver LH, Grütter MG, and Matthews BW. (1995). The refined structures of goose lysozyme and its complex with a bound trisaccharide show that the "goose-type" lysozymes lack a catalytic aspartate residue. J Mol Biol. 1995;245(1):54-68. DOI:10.1016/s0022-2836(95)80038-7 |
- Xiang Y, Morais MC, Cohen DN, Bowman VD, Anderson DL, and Rossmann MG. (2008). Crystal and cryoEM structural studies of a cell wall degrading enzyme in the bacteriophage phi29 tail. Proc Natl Acad Sci U S A. 2008;105(28):9552-7. DOI:10.1073/pnas.0803787105 |